Search Count: 45
 |
Organism: Escherichia coli dsm 30083 = jcm 1649 = atcc 11775
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2025-06-04 Classification: TOXIN |
 |
Organism: Homo sapiens, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2025-02-12 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-02-12 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-02-12 Classification: PROTEIN BINDING |
 |
Structure Of Human Argonaute2-Guide-Target Complex In A Fully Paired, Slicing-Competent Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2024-12-11 Classification: HYDROLASE/RNA Ligands: MG |
 |
Ternary Complex Structure Of Cereblon-Ddb1 Bound To Wiz(Zf7) And The Molecular Glue Wiz-5
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2024-11-27 Classification: LIGASE Ligands: ZN, SO4, A1A5H |
 |
Ternary Complex Structure Of Cereblon-Ddb1 Bound To Wiz(Zf7) And The Molecular Glue Wiz-6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2024-11-27 Classification: LIGASE Ligands: ZN, SO4, A1A5I |
 |
Ternary Complex Structure Of Cereblon-Ddb1 Bound To Wiz(Zf7) And The Molecular Glue Dwiz-1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-07-10 Classification: LIGASE Ligands: ZN, U3I, SO4, EDO |
 |
Organism: Bluetongue virus (serotype 1 / isolate south africa)
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: VIRAL PROTEIN |
 |
Organism: Bluetongue virus (serotype 1 / isolate south africa)
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: VIRAL PROTEIN |
 |
Organism: Bluetongue virus (serotype 1 / isolate south africa)
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: VIRAL PROTEIN |
 |
Organism: Bluetongue virus (serotype 1 / isolate south africa)
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: VIRAL PROTEIN |
 |
Organism: Bluetongue virus (serotype 1 / isolate south africa)
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: VIRAL PROTEIN |
 |
Organism: Bluetongue virus (serotype 1 / isolate south africa)
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: VIRAL PROTEIN |
 |
Organism: Bluetongue virus (serotype 1 / isolate south africa)
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: VIRAL PROTEIN |
 |
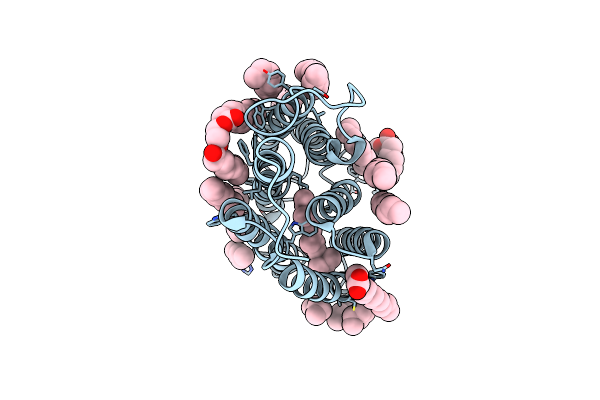
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.56 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
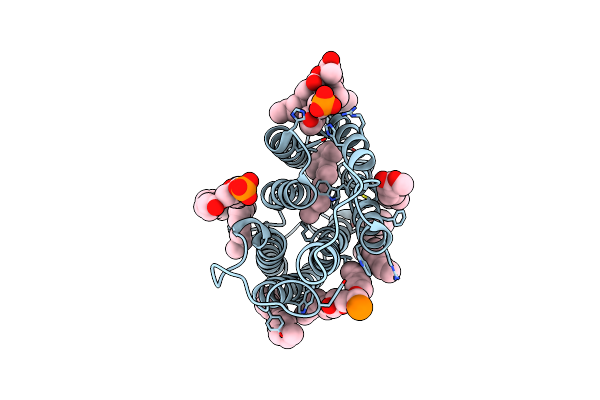
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr2 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.53 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
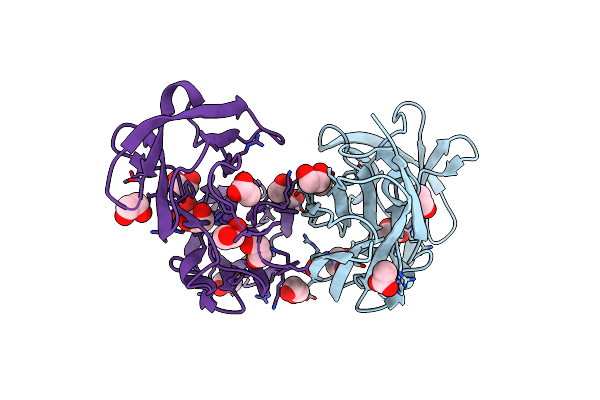
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 H225F Mutant In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Organism: Scylla paramamosain
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-10-26 Classification: ALLERGEN Ligands: EDO, ACT |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-23 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |

