Search Count: 83
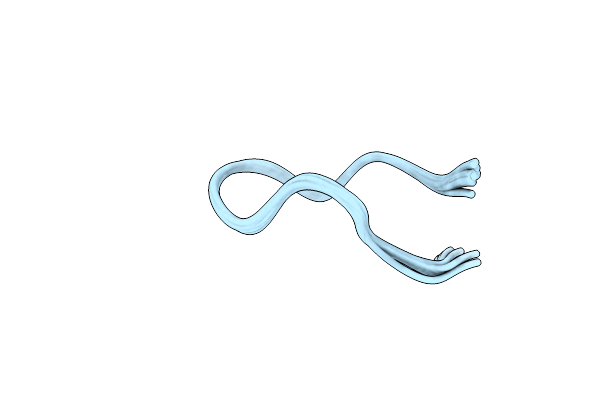 |
Structure Of The Globular Isoform Of The Novel Conotoxin Pnid Derived From Conus Pennaceus
|
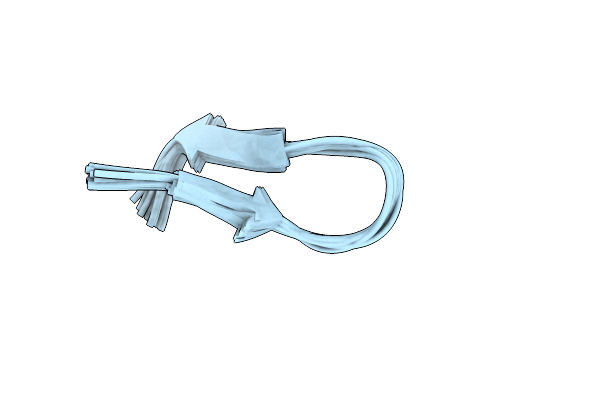 |
Structure Of The Ribbon Isoform Of The Novel Conotoxin Pnid Derived From Conus Pennaceus
|
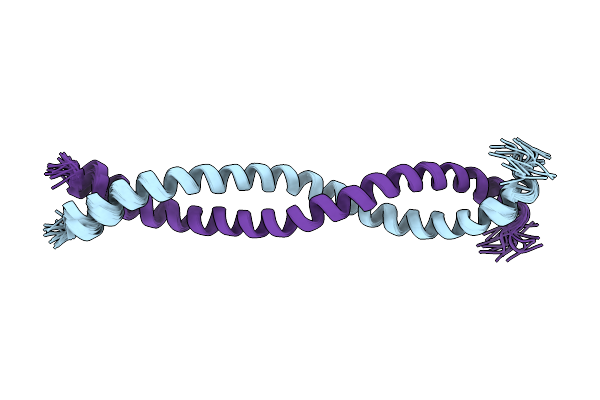 |
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2021-03-24 Classification: RNA BINDING PROTEIN |
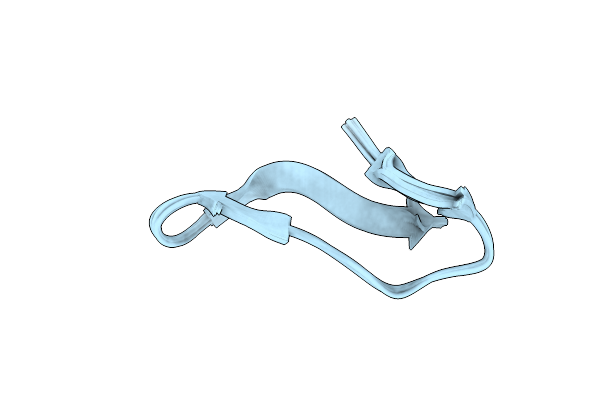 |
Structural Characterization Of Novel Conotoxin Miiib Derived From Conus Magus
|
 |
N-Terminal Domain Of Dynein Intermediate Chain From Chaetomium Thermophilum
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: SOLUTION NMR Release Date: 2020-08-19 Classification: MOTOR PROTEIN |
 |
X-Ray Structure Of The Pglf Dehydratase From Campylobacter Jejuni In Complex With Udp And Nad(H)
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-11-08 Classification: MEMBRANE PROTEIN Ligands: UDP, NAD, EDO, NA |
 |
X-Ray Structure Of The Pglf Udp-N-Acetylglucosamine 4,6-Dehydratase From Campylobacterjejuni, D396N/K397A Variant In Complex With Udp-N-Acrtylglucosamine
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-11-08 Classification: MEMBRANE PROTEIN Ligands: NAD, UD1, EDO, NA |
 |
X-Ray Structure Of The Pglf 4,6-Dehydratase From Campylobacter Jejuni, T595S Variant, In Complex With Udp
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-11-08 Classification: MEMBRANE PROTEIN Ligands: UDP, NAD, EDO, NA |
 |
X-Ray Structure Of The Pglf 4,6-Dehydratase From Campylobacter Jejuni, Variant T395V, In Complex With Udp
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-11-08 Classification: MEMBRANE PROTEIN Ligands: UDP, NAD, EDO, NA |
 |
X-Ray Structure Of The Pglf 4,5-Dehydratase From Campylobacter Jejuni, Variant M405Y, In Complex With Udp
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-11-08 Classification: MEMBRANE PROTEIN Ligands: UDP, NAD, EDO, NA |
 |
Organism: Wisteria floribunda
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2016-09-14 Classification: SUGAR BINDING PROTEIN Ligands: NGA, CA, MN |
 |
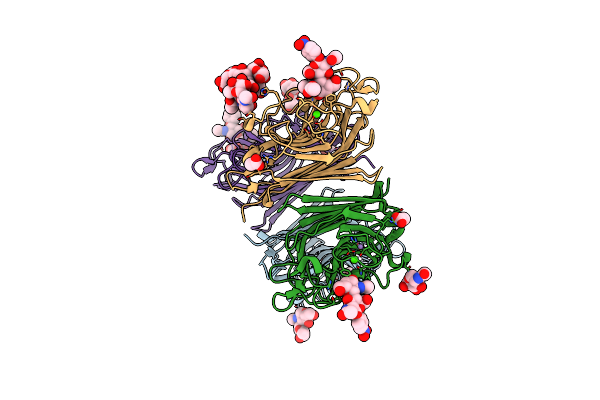
Wisteria Floribunda Lectin In Complex With Galnac(Beta1-4)Glcnac (Lacdinac) At Ph 8.5.
Organism: Wisteria floribunda
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-14 Classification: SUGAR BINDING PROTEIN Ligands: 6Y2, CA, MN, NAG |
 |
Wisteria Floribunda Lectin In Complex With Galnac(Beta1-4)Glcnac (Lacdinac) At Ph 6.5
Organism: Wisteria floribunda
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-09-14 Classification: SUGAR BINDING PROTEIN Ligands: CA, MN, ACT, 6Y2, NAG |
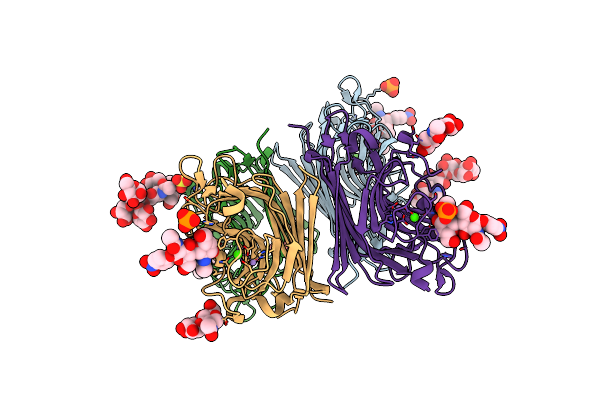 |
Wisteria Floribunda Lectin In Complex With Galnac(Beta1-4)Glcnac (Lacdinac) At Ph 4.2
Organism: Wisteria floribunda
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2016-09-14 Classification: sugar binding protein/inhibitor Ligands: 6Y2, MN, CA, PO4, SO4 |
 |
High Resolution Solution Nmr Structure Of The Spider Venom Peptide U3- Scytotoxin-Sth1A
|
 |
High Resolution Solution Nmr Structure Of The Spider Venom Peptide U3- Scytotoxin-Sth1H
|
 |
High Resolution Solution Nmr Structure Of The Spider Venom Peptide U5- Scytotoxin-Sth1A
|
 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-07-29 Classification: TRANSFERASE Ligands: 4RA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-11-27 Classification: HYDROLASE Ligands: ZN, EPE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2013-11-06 Classification: HYDROLASE Ligands: ZN |

