Search Count: 4,754
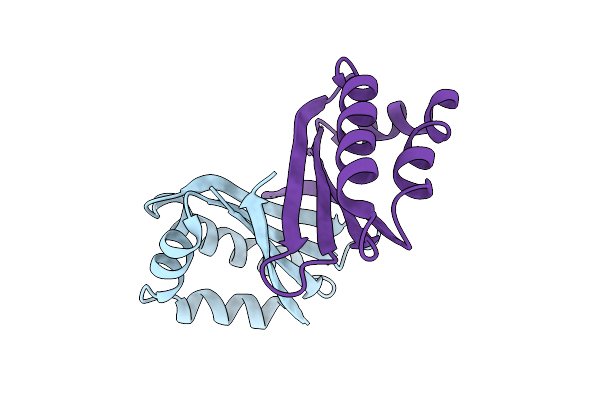 |
An Antibiotic Biosynthesis Monooxygenase Family Protein From Streptomyces Sp. Ma37
Organism: Streptomyces sp. ma37
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
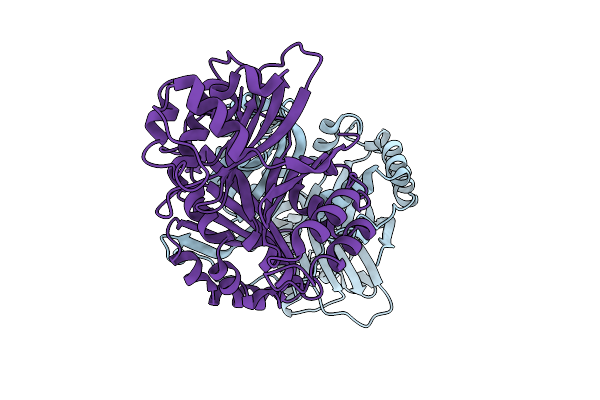 |
Crystal Structure Of A Polyketide Abm/Scha-Like Domain-Containing Protein Whie-Orfi From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
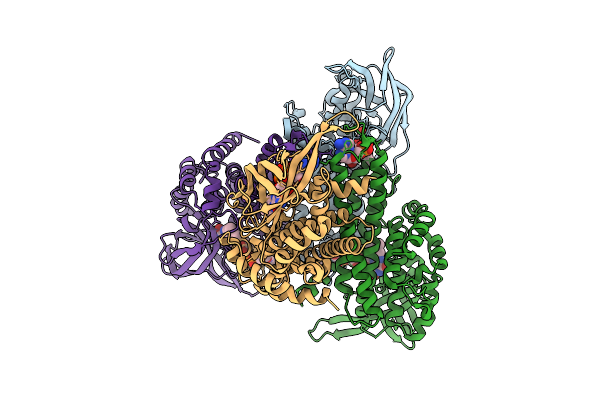 |
Organism: Faecalibacterium prausnitzii l2-6
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: FAD |
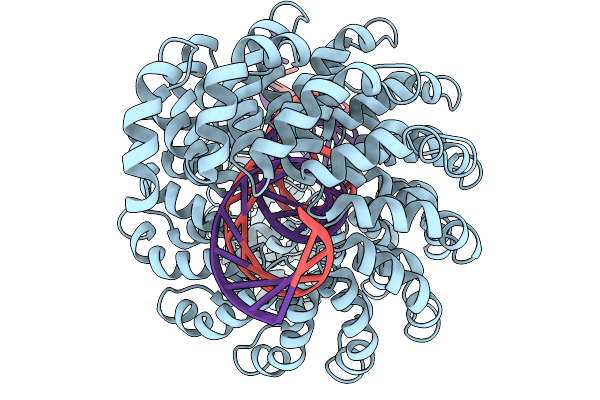 |
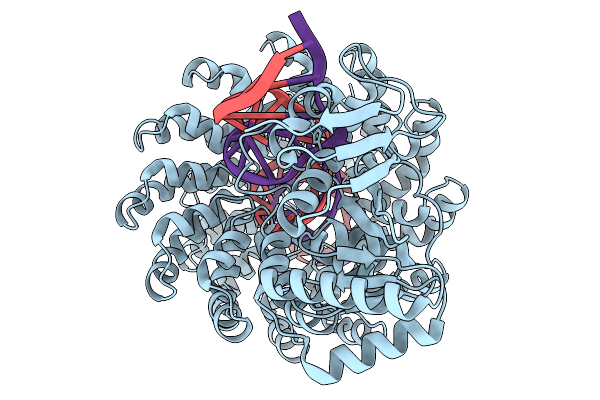
Organism: Burkholderia cenocepacia, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
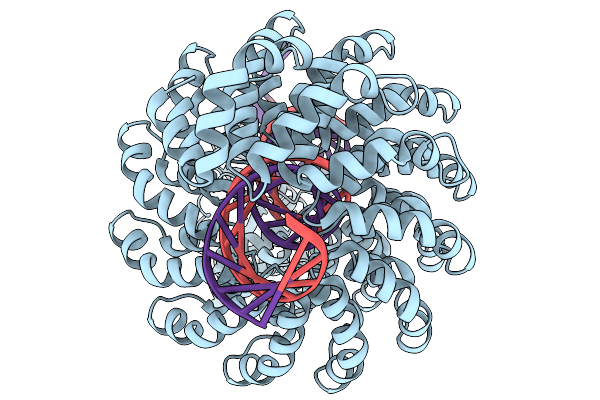
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
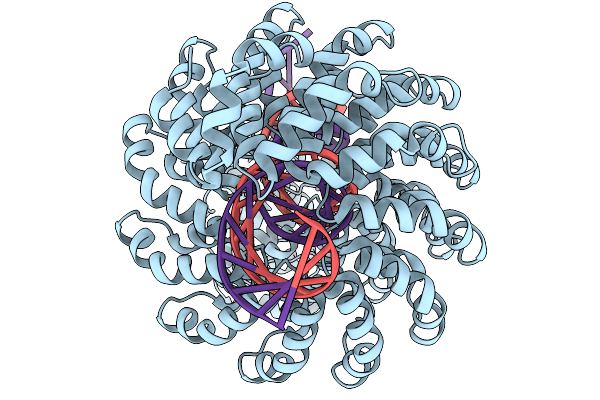
Organism: Xanthomonas, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
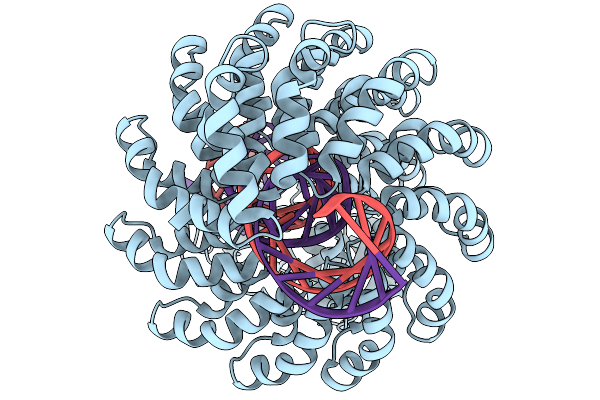
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
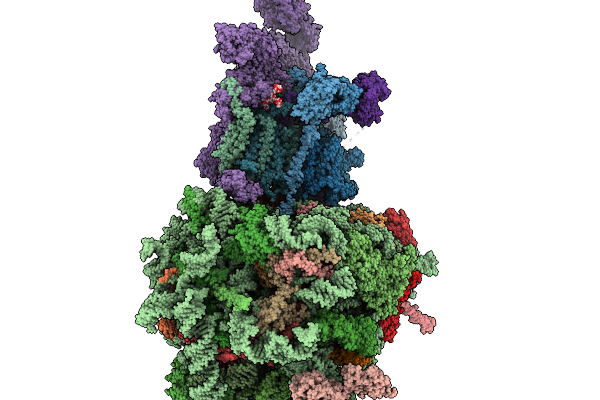 |
Structure Of A Grp94 Folding Intermediate Engaged With A Ccdc134- And Fkbp11-Bound Secretory Translocon
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: RIBOSOME Ligands: ELU, MG, ZN |
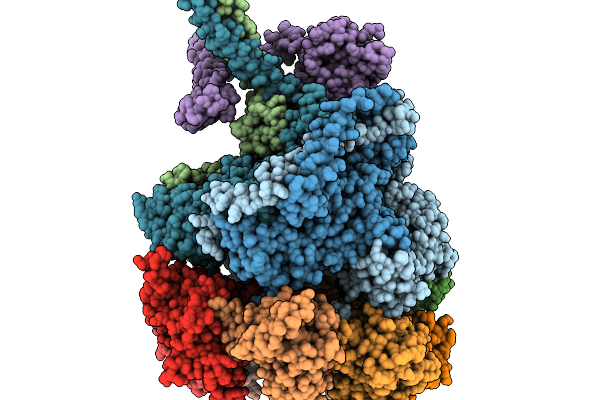 |
Nub1/Fat10-Processing Human 26S Proteasome With Rpt2 At Top Of Spiral Staircase And Partially Unfolded Eos (Aaa+ Motor Locally Refined)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE/PROTEIN BINDING Ligands: ATP, MG, ADP, ZN |
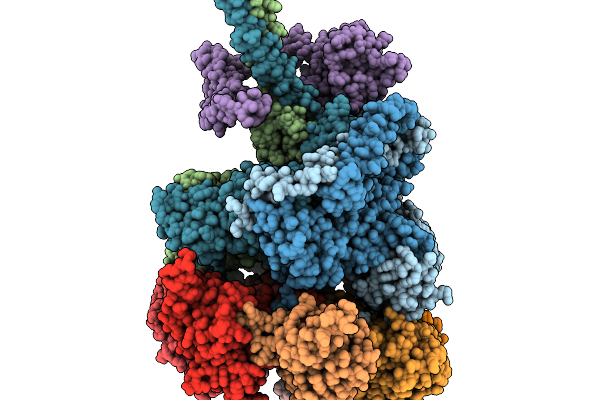 |
Nub1/Fat10-Processing Human 26S Proteasome With Rpt5 At Top Of Spiral Staircase (Aaa+ Locally Refined)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MOTOR PROTEIN Ligands: ADP, MG, ZN, ATP |
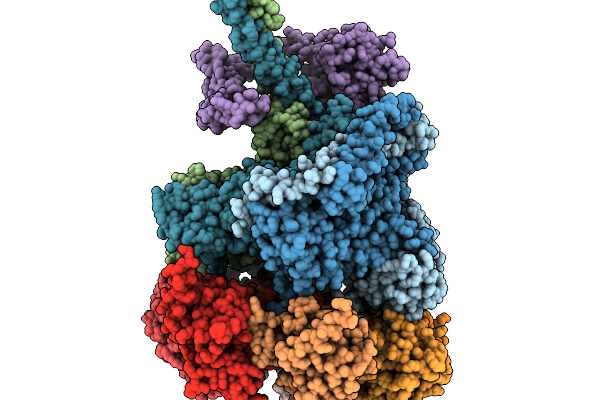 |
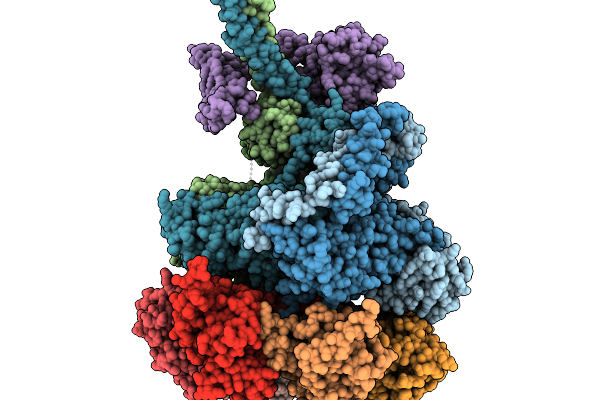
Nub1/Fat10-Processing Human 26S Proteasome With Rpt1 At Top Of Spiral Staircase (Aaa+ Locally Refined)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MOTOR PROTEIN Ligands: ATP, MG, ADP, ZN |
 |
Nub1/Fat10-Processing Human 26S Proteasome With Rpt4 At Top Of Spiral Staircase (Aaa+ Motor Locally Refined)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MOTOR PROTEIN Ligands: ADP, ZN, ATP, MG |
 |
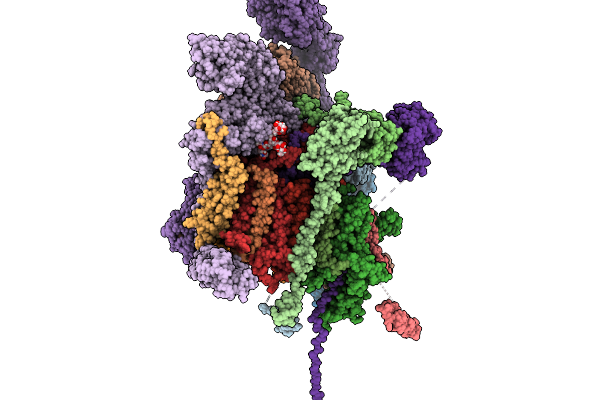
Cryo-Em Structure Of The Type Ii Secretion System Protein From Acidithiobacillus Caldus
Organism: Acidithiobacillus caldus (strain sm-1)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TOXIN |
 |
Structure Of A Grp94 Folding Intermediate Engaged With A Ccdc134- And Fkbp11-Bound Secretory Translocon
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: RIBOSOME Ligands: ELU |
 |
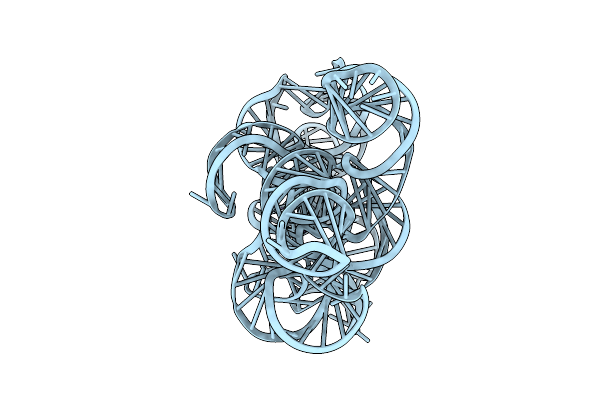
Cryo-Em Structure Of Linear Intron Of Thymidylate Synthase (Td) Gene Of Bacteriophage T4
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA Ligands: MG |
 |
Cryo-Em Structure Of Circular Intron Of Thymidylate Synthase (Td) Gene Of Bacteriophage T4
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA Ligands: MG |
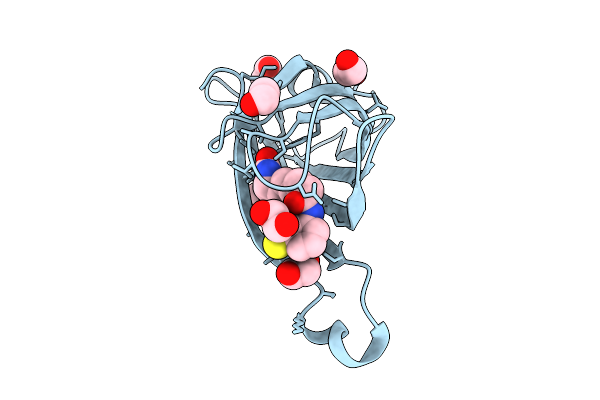 |
Streptavidin With A Thiophenol Cofactor As Artificial Hydrogen Atom Transferase
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSFERASE Ligands: A1I7Y, EDO |
 |
Streptavidin K121W With A Thiophenol Cofactor As Artificial Hydrogen Atom Transferase
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSFERASE Ligands: A1I7Y |
 |
Streptavidin 112Y-121W-124F With A Thiophenol Cofactor As Artificial Hydrogen Atom Transferase
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSFERASE Ligands: A1I7Y, ACT, EDO |

