Search Count: 2,168
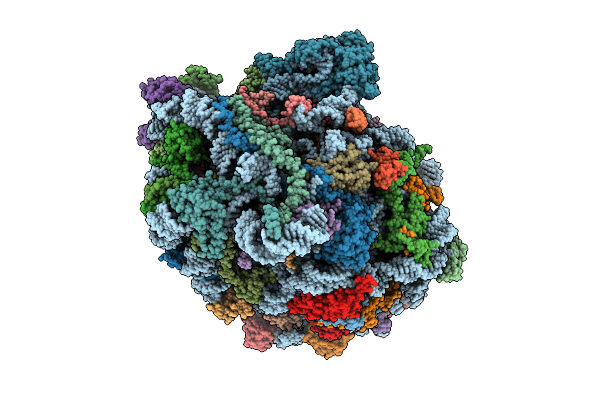 |
High Resolution Structure Of The Thermophilic 60S Ribosomal Subunit Of Chaetomium Thermophilum
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, SPM, MG, K, ZN |
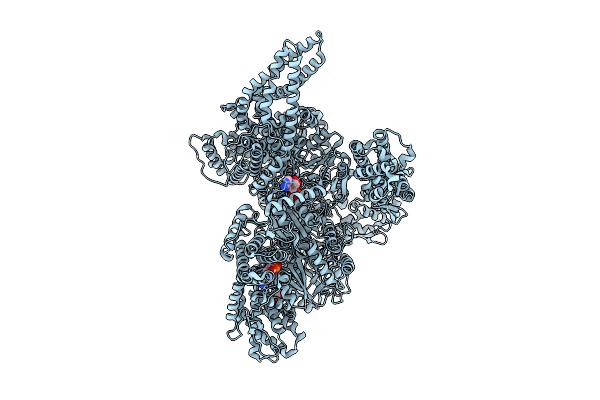 |
Motor Domain With Apo Aaa1 And Adp Aaa3 From Yeast Full-Length Dynein-1 In 0.1 Mm Atp Condition
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MOTOR PROTEIN Ligands: ATP, ADP, MG |
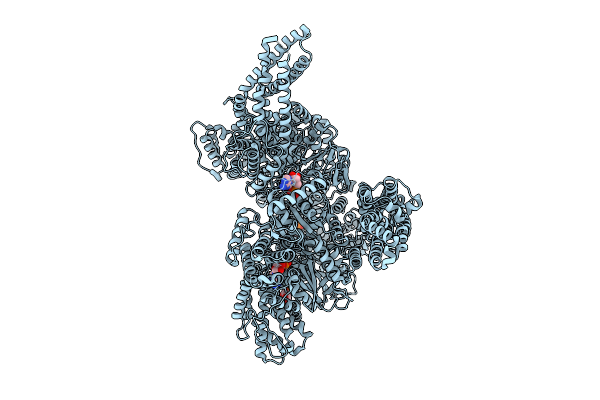 |
Motor Domain With Adp Aaa1 And Adp Aaa3 From Yeast Full-Length Dynein-1 In 0.1 Mm Atp Condition
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MOTOR PROTEIN Ligands: ADP, ATP |
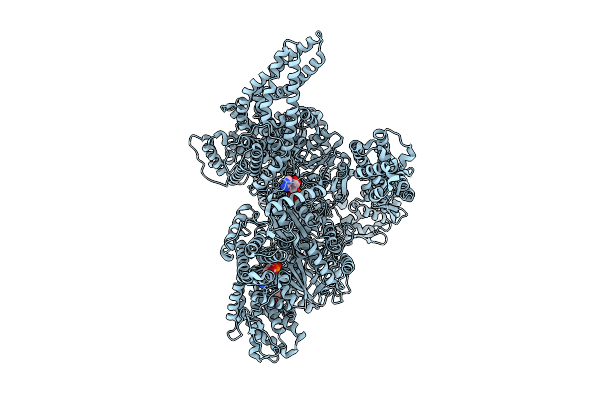 |
Motor Domain Alone With Apo Aaa1 And Adp Aaa3 From Yeast Full-Length Dynein-1 And Pac1 In 0.1 Mm Atp Condition
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MOTOR PROTEIN Ligands: ATP, ADP, MG |
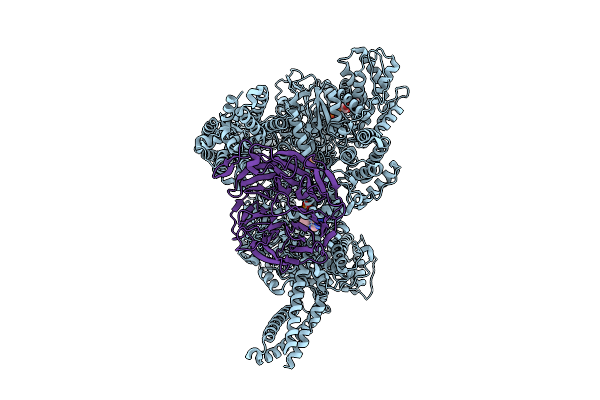 |
Motor Domain-Pac1 Complex With Adp Aaa1 And Apo Aaa3 From Yeast Full-Length Dynein-1 And Pac1 In 0.1 Mm Atp Condition
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MOTOR PROTEIN Ligands: ADP, ATP, MG |
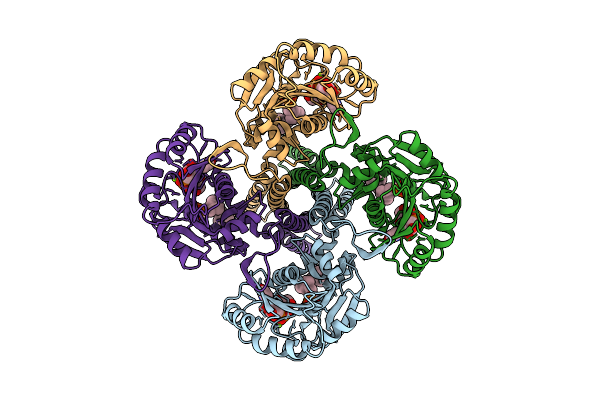 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Substrate-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
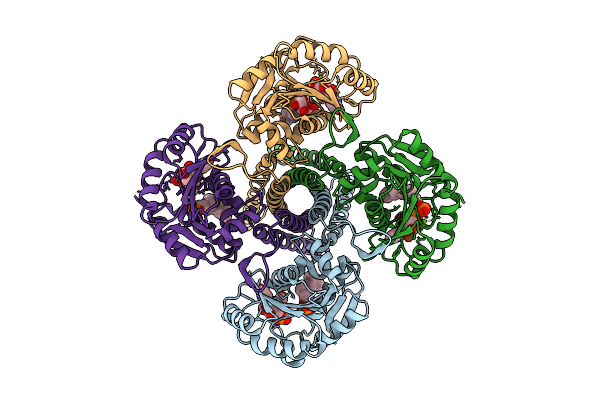 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Catalysis And Product-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG, UDP, A1B94 |
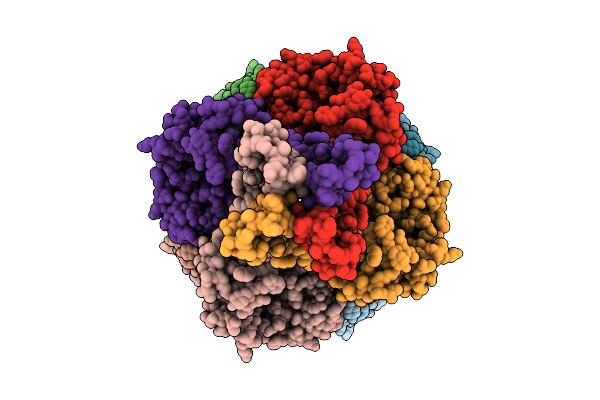 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Apo State (Octamer Volume)
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Intermediate State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Structure Of The Saccharomyces Cerevisiae Pmt4-Mir Domain With Bound Ligands
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: PEPTIDE BINDING PROTEIN Ligands: EPE, GOL |
 |
Structure Of The Chaetomium Thermophilum Pmt4-Mir Domain With Bound Ligands
Organism: Chaetomium
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: PEPTIDE BINDING PROTEIN Ligands: EDO, ACT, NA |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |

