Search Count: 95
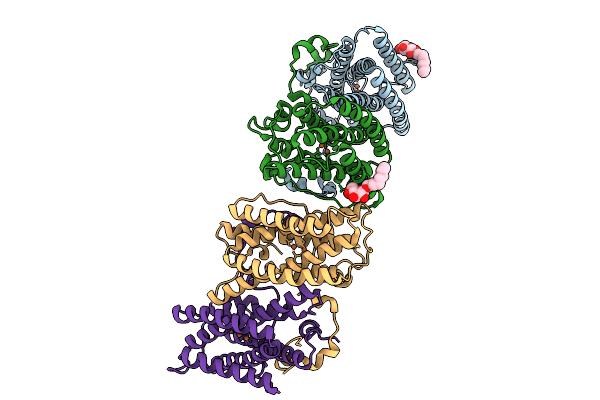 |
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: OXIDOREDUCTASE Ligands: FE, OH, BOG |
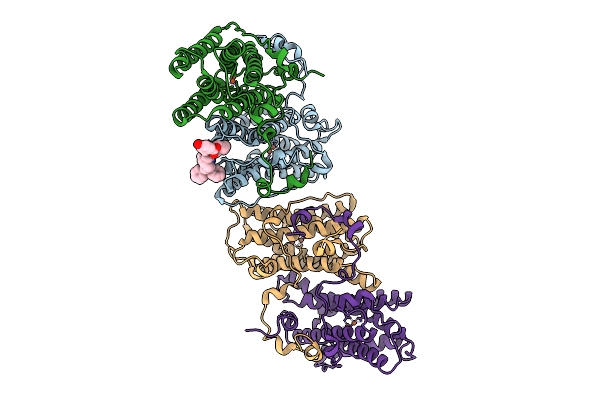 |
The Crystal Structure Of The Trypanosome Alternative Oxidase Complexed With A Trypanocidal Phosphonium Derivative (Compound1)
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: OXIDOREDUCTASE Ligands: FE, OH, A1L8E |
 |
Organism: Trypanosoma brucei brucei
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: MEMBRANE PROTEIN Ligands: VO6 |
 |
Organism: Trypanosoma brucei brucei
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: MEMBRANE PROTEIN Ligands: PNT |
 |
Organism: Trypanosoma brucei brucei
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: MEMBRANE PROTEIN Ligands: GOL |
 |
Crystal Structure Of Human Eif2 Alpha-Gamma Complexed With Ppp1R15A_420-452
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2024-03-20 Classification: TRANSLATION Ligands: GNP |
 |
Organism: Homo sapiens, Oryctolagus cuniculus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-09-29 Classification: HYDROLASE Ligands: ATP, CA |
 |
Cryo-Em Structure Of Pre-Dephosphorylation Complex Of Phosphorylated Eif2Alpha With Trapped Holophosphatase (Pp1A_D64A/Ppp1R15A/G-Actin/Dnase I)
Organism: Homo sapiens, Oryctolagus cuniculus, Escherichia coli (strain k12), Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: HYDROLASE Ligands: MN, ATP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: NAD, MG, FUM, TTN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: NAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: NAD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSLATION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSLATION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSLATION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSLATION |
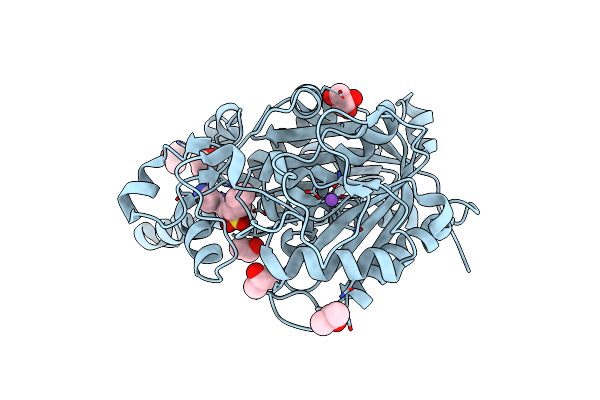 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-04-01 Classification: TRANSFERASE |
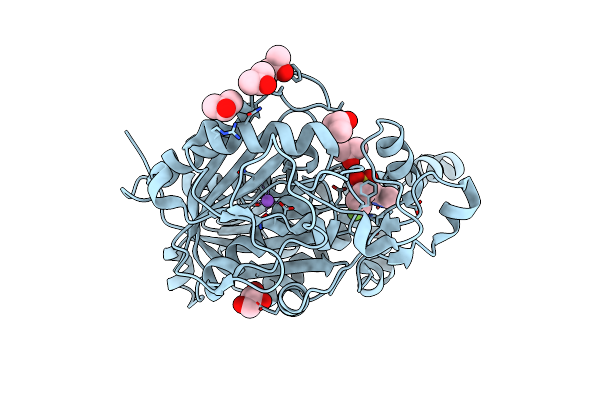 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2020-04-01 Classification: TRANSFERASE Ligands: JFX, NA, GOL, IPA |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2020-04-01 Classification: TRANSFERASE |
 |
The Crystal Structure Of Phytoplasmal Effector Causing Phyllody Symptoms 1 (Phyl1)
Organism: 'echinacea purpurea' witches'-broom phytoplasma
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2019-07-31 Classification: GENE REGULATION Ligands: CD |
 |
Crystal Structure Of Human Phosphodiesterase 4D2 Catalytic Domain With Inhibitor Npd-048
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2019-03-20 Classification: HYDROLASE Ligands: ZN, MG, EDO, EPE, E6Z |

