Search Count: 285
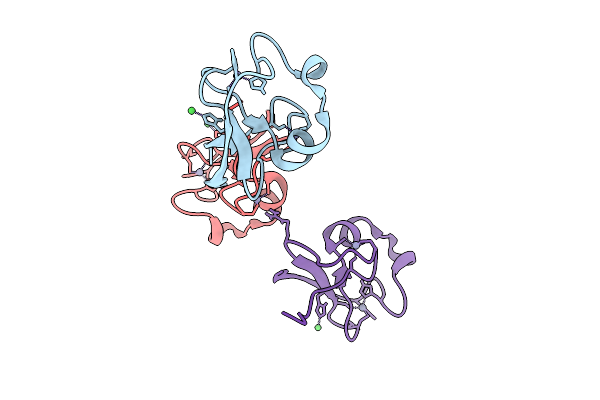 |
R-Degron Fused Zz-Domain Of The Arabidopsis Thaliana E3 Ubiquitin-Protein Ligase Big
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: LIGASE Ligands: ZN, NI |
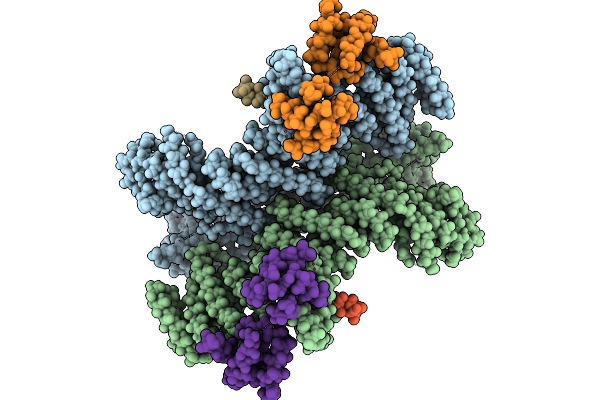 |
Cryo-Em Structure Of The Core Of The Arabidopsis Thaliana Ubr4/Di19/Calm1 Complex
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
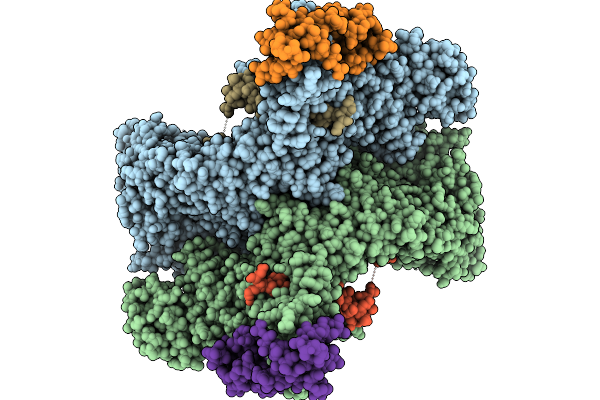 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (C-Term Dimer Interface Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
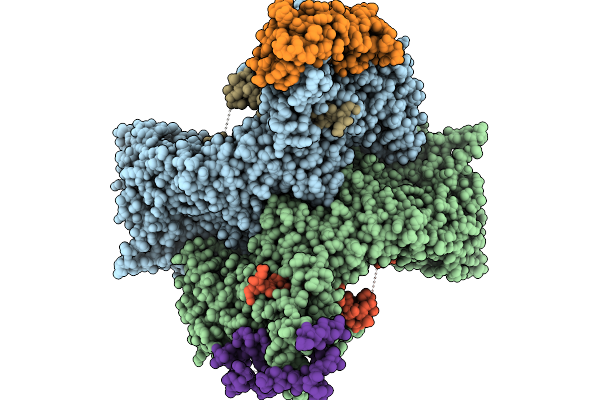 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (Calm1 Focused Refinement)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
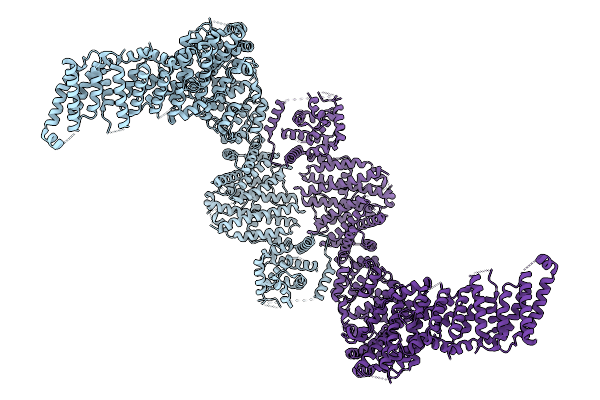 |
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (N-Term Focused Refinement)
|
 |
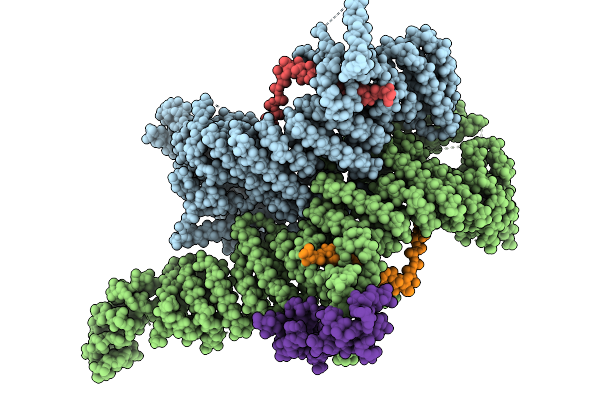
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (Bp Focused Refinement)
|
 |
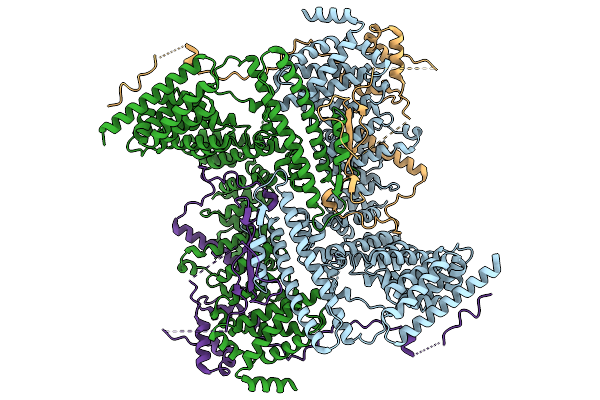
Cryo-Em Structure Of The Human Ubr4/Kcmf1/Calm1 Complex (C-Term Focused Refinement)
|
 |
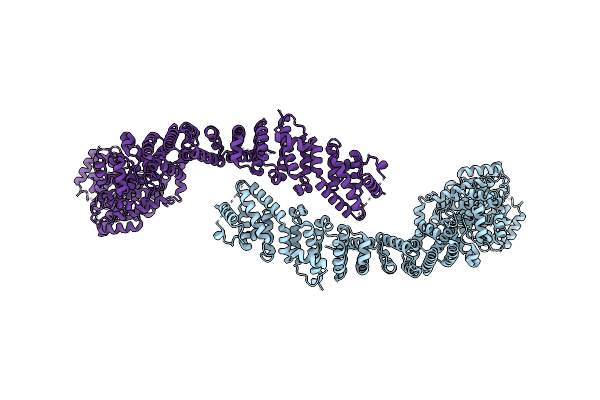
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (C-Term Dimer Interface Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE Ligands: ZN |
 |
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (N-Term Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
 |
Cryo-Em Structure Of The C. Elegans Ubr4/Kcmf1 Complex (Side Focused Refinement)
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: LIGASE Ligands: ZN |
 |
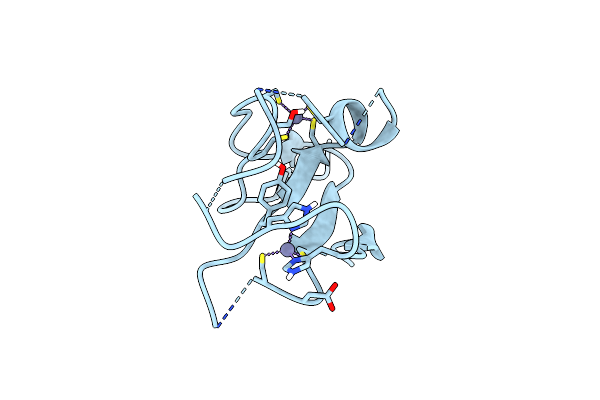
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2025-06-04 Classification: LIGASE Ligands: ZN, PO4, K |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-05-14 Classification: LIGASE Ligands: ZN |
 |
Y-Degron Fused Zz-Domain Of The Arabidopsis Thaliana E3 Ubiquitin-Protein Ligase Prt1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-05-14 Classification: LIGASE Ligands: ZN, MG |
 |
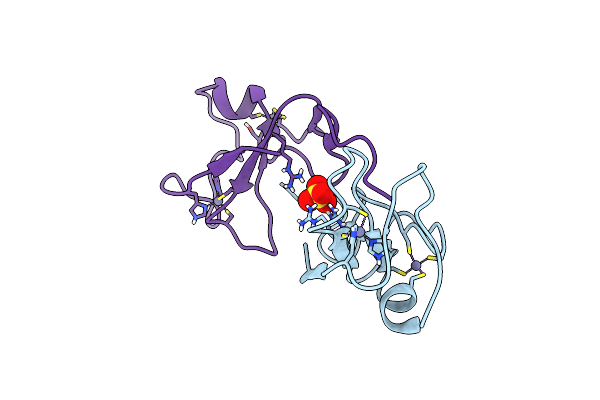
F-Degron Fused Zz-Domain Of The Arabidopsis Thaliana E3 Ubiquitin-Protein Ligase Prt1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-05-14 Classification: LIGASE Ligands: ZN, MG |
 |
W-Degron Fused Zz-Domain Of The Arabidopsis Thaliana E3 Ubiquitin-Protein Ligase Prt1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2025-05-14 Classification: LIGASE Ligands: ZN, SO4 |
 |
High-Resolution Ambient Temperature Structure Of Lysozyme Soaked With Sodium Iodide
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: GOL, NA, IOD |
 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: CA |
 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin And A Nicotinamide Adenine Dinucleotide (Nad+)
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: NAD, CA, GOL |

