Search Count: 44
 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE Ligands: NCD, ZN |
 |
Cryo-Em Structure Of Lptdem Complex Containing Shigella Flexneri Lpte And Endogenous E. Coli Lptd And Lptm
Organism: Shigella flexneri, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLM, PXS |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde In Complex With Rtp45 Superinfection Exclusion Protein From Rtp Bacteriophage And Endogenous Lptm
Organism: Shigella flexneri, Escherichia phage rtp, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLM, PXS |
 |
Cryo-Em Structure Of The Open State Of Shigella Flexneri Lptde Bound By The Rbp Of Oekolampad Phage
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
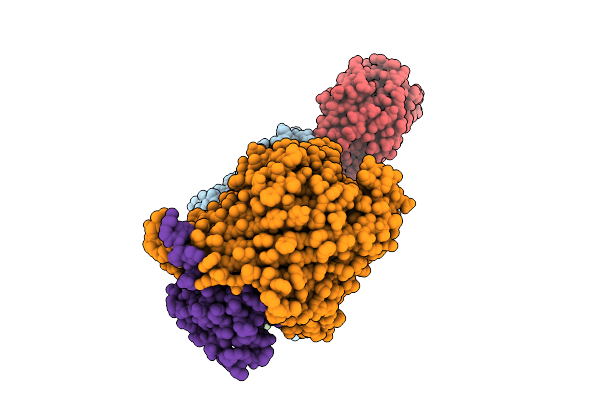 |
Cryo-Em Structure Of Shigella Flexneri Lptde Dimer: Closed-State Unbound And Open-State Bound By Oekolampad Phage Rbp
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
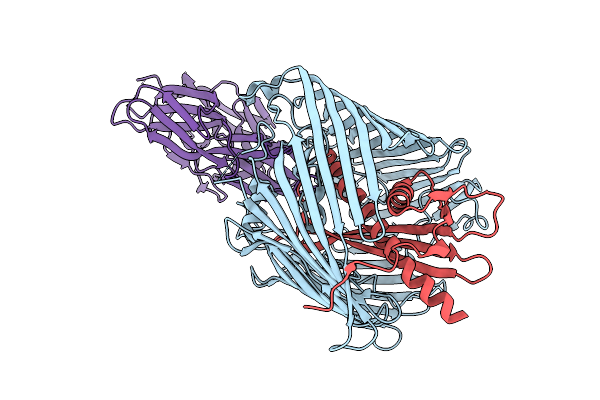 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By Phage Rbp Reveals N-Terminal Strand Insertion Into Lateral Gate
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
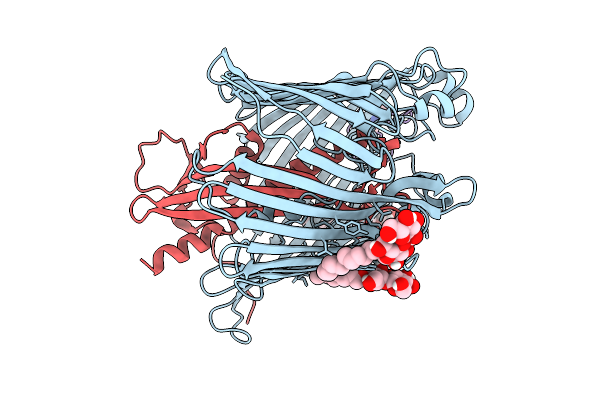
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 1)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, A1I1D |
 |
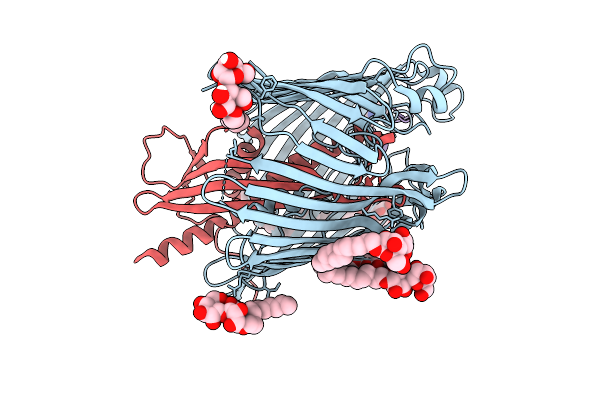
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 2)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, KZ0 |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 3)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, KZ0 |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 4)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, A1I1E |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 5)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, R06 |
 |
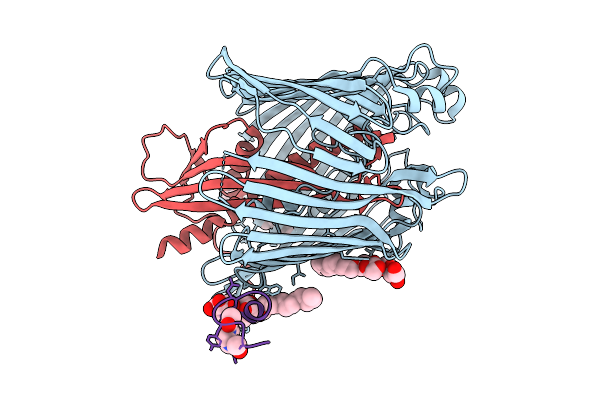
Cryo-Em Structure Of Shigella Flexneri Lptde In Complex With A Bicyclic Peptide Binder (Compound 12)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, A1I1D |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 13)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, R06 |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By A Bicyclic Peptide Molecule (Compound 16)
Organism: Shigella flexneri, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: LMT, PLM, Z41, A1I4O |
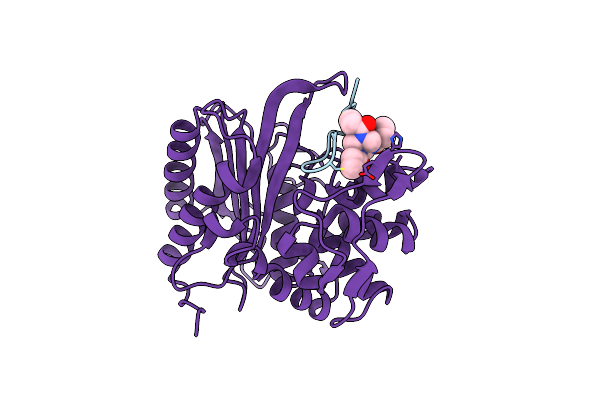 |
The Structure Of E. Coli Penicillin Binding Protein 3 (Pbp3) In Complex With A Bicyclic Peptide Inhibitor
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-04-03 Classification: HYDROLASE Ligands: 29N |
 |
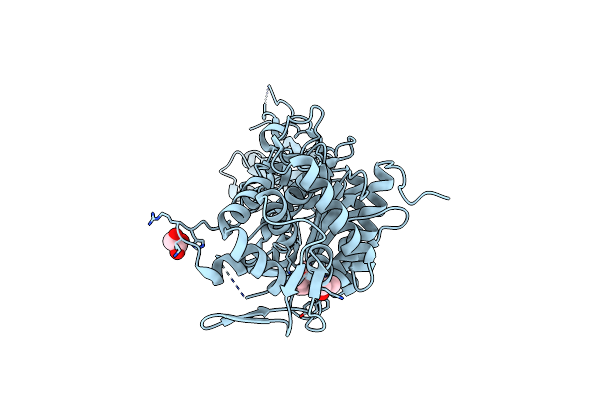
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: ZN |
 |
Structure Of P. Aeruginosa Pbp3 In Complex With A Phenyl Boronic Acid (Compound 1)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-08-11 Classification: HYDROLASE Ligands: GOL, RY2, DMS |
 |
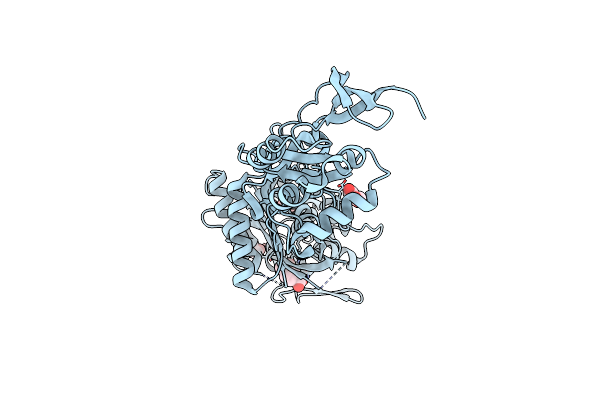
Structure Of P. Aeruginosa Pbp3 In Complex With An Aryl Boronic Acid (Compound 2)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2021-08-11 Classification: HYDROLASE Ligands: RXW, DMS, GOL |
 |
Structure Of P. Aeruginosa Pbp3 In Complex With A Benzoxaborole (Compound 3)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2021-08-11 Classification: HYDROLASE Ligands: GOL, RZZ |
 |
Structure Of P. Aeruginosa Pbp3 In Complex With A Benzoxaborole (Compound 4)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-08-11 Classification: HYDROLASE Ligands: S1B |

