Search Count: 116
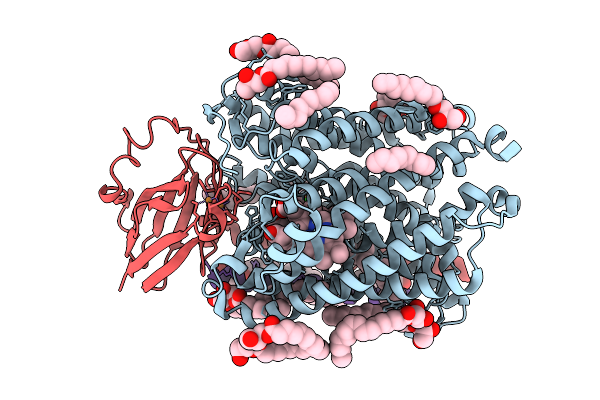 |
Chloride Bound Structure Of Oxidized Ba3-Type Cytochrome C Oxidase Confirmed By Single-Wavelength Anomalous Diffraction
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: CU, HEM, HAS, OLC, CL, CUA |
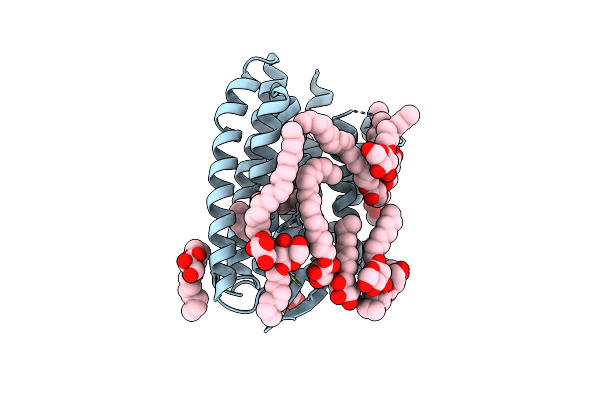 |
Grouped 150-240 Ms Dark Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL, BOG, MPG |
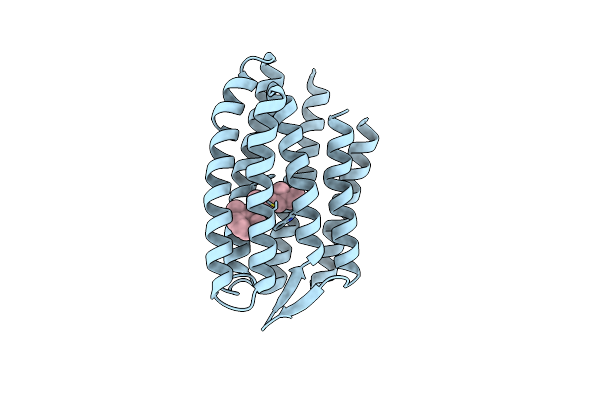 |
Continuously Illuminated Structure Of Sensory Rhodopsin Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL |
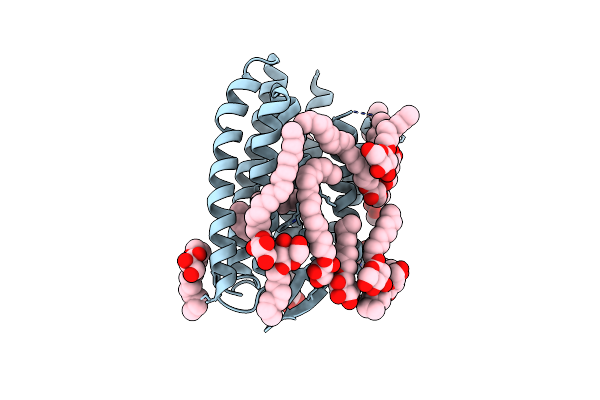 |
Continuous Dark State Structure Of Sensory Rhodopsin Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL, BOG, MPG |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 90-120 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 60-90 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography. 30-60 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 0-30 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 120-150 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Laser Excitation Effects On Br: Reprocessed Dark Dataset Recorded From Nango Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Reprocessed Extrapolated 16 Ns Light Dataset Recorded At 0.04 Gw/Cm2 From Nango Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 6 Ps Light Dataset Recorded At 342 Gw/Cm2 At Swissfel
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 6 Ps Light Dataset Recorded At 2493 Gw/Cm2 At Swissfel
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Reprocessed Extrapolated 10Ps Light Dataset Recorded At 525 Gw/Cm2 From Nogly Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 10Ps Light Dataset Recorded At 1281 Gw/Cm2 At Sacla (+100Ps, 1Ns, 10Ns)
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Room-Temperature Serial Synchrotron Crystallography Structure Of Spinacia Oleracea Rubisco
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-05-29 Classification: LYASE Ligands: MG |
 |
Dark State Structure Of Sensory Rhodopsin Ii In Complex With Htrii Solved By Room Temperature Serial Synchrotron Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-11-29 Classification: MEMBRANE PROTEIN Ligands: RET |

