Search Count: 19
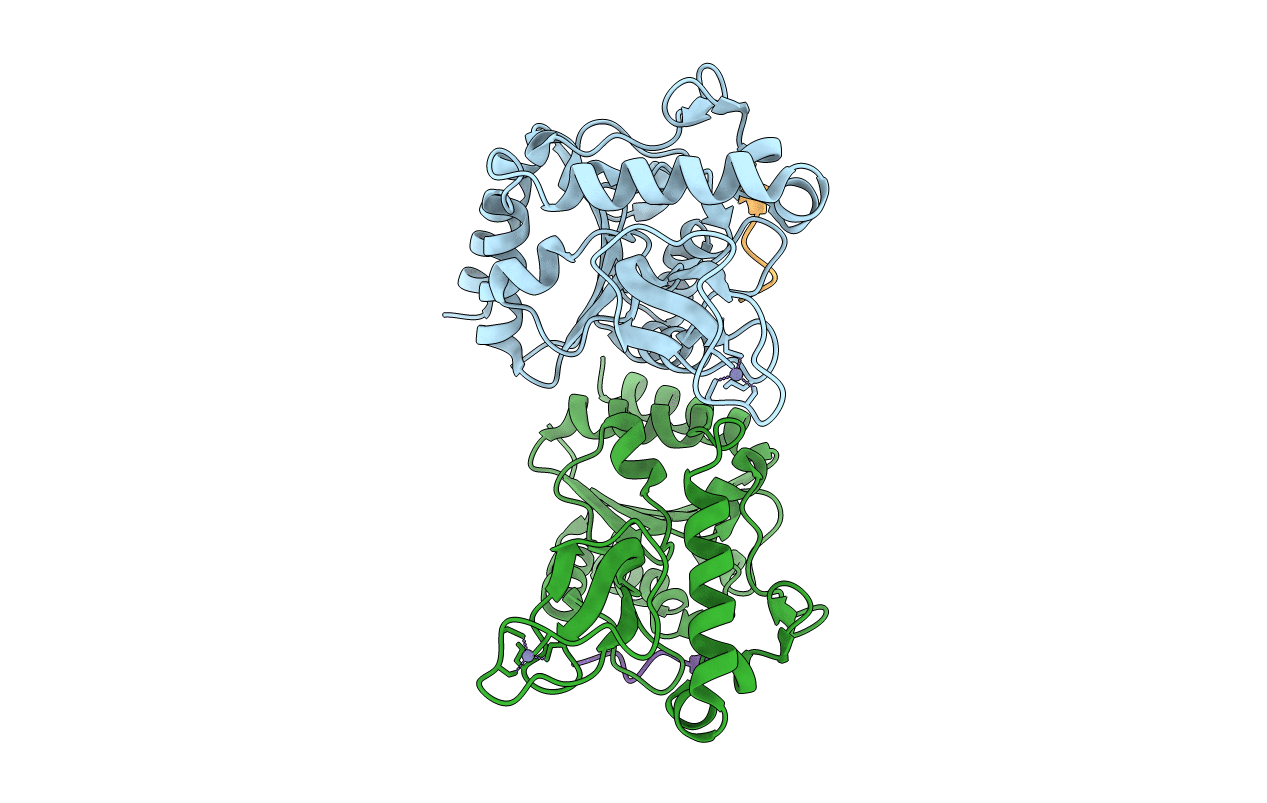 |
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: ZN |
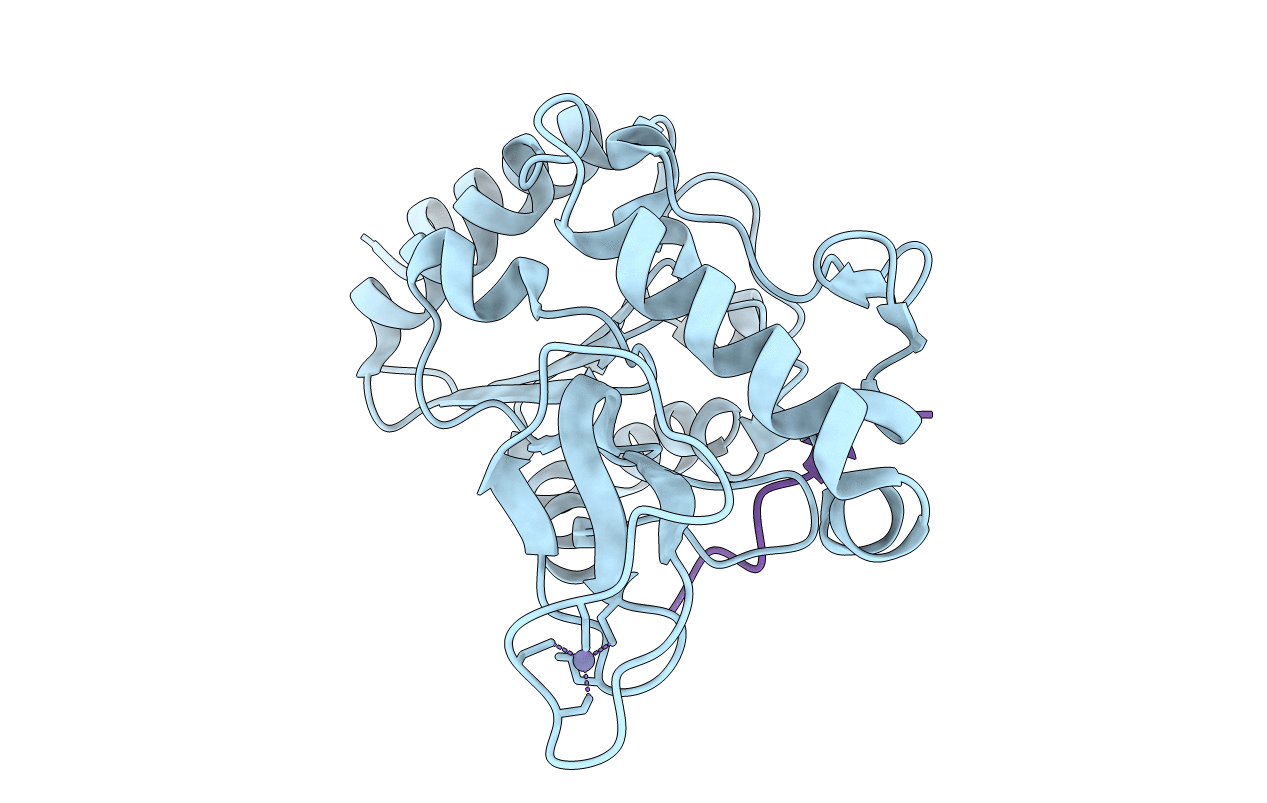 |
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: ZN |
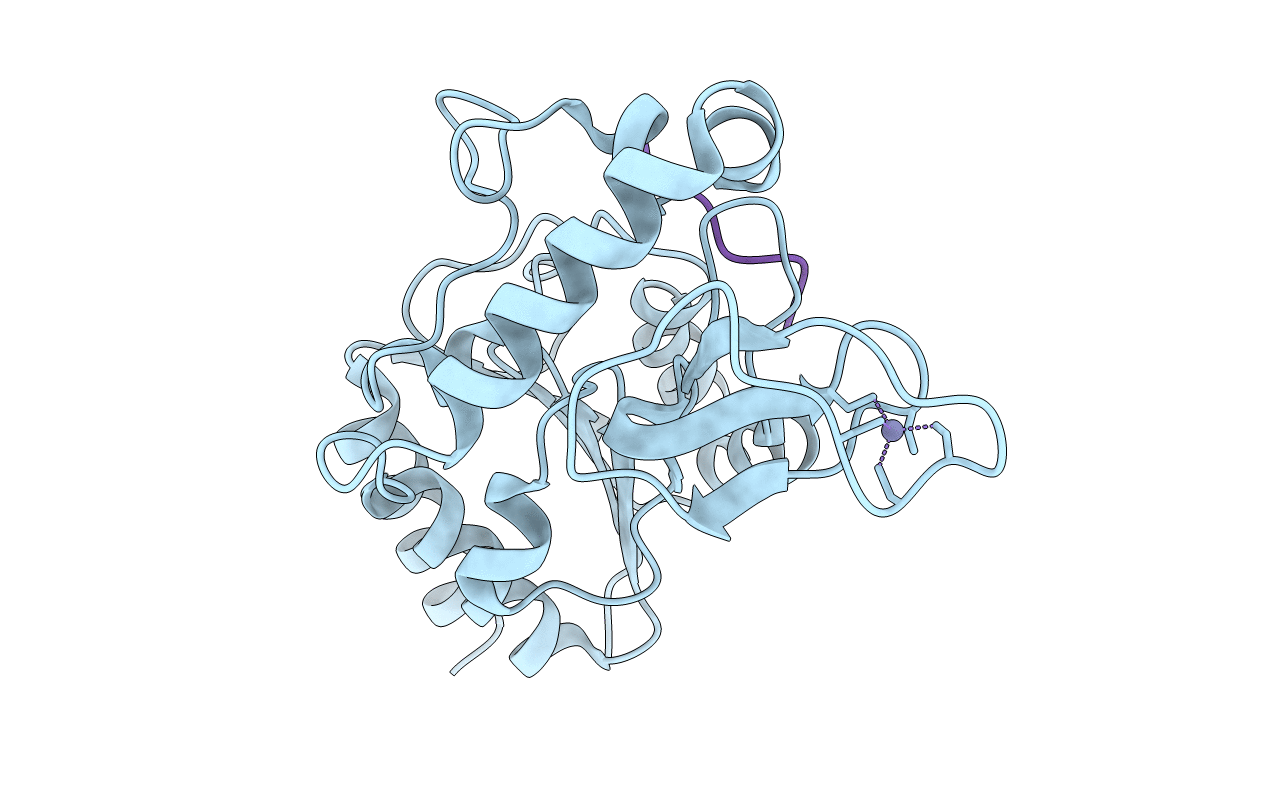 |
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: ZN |
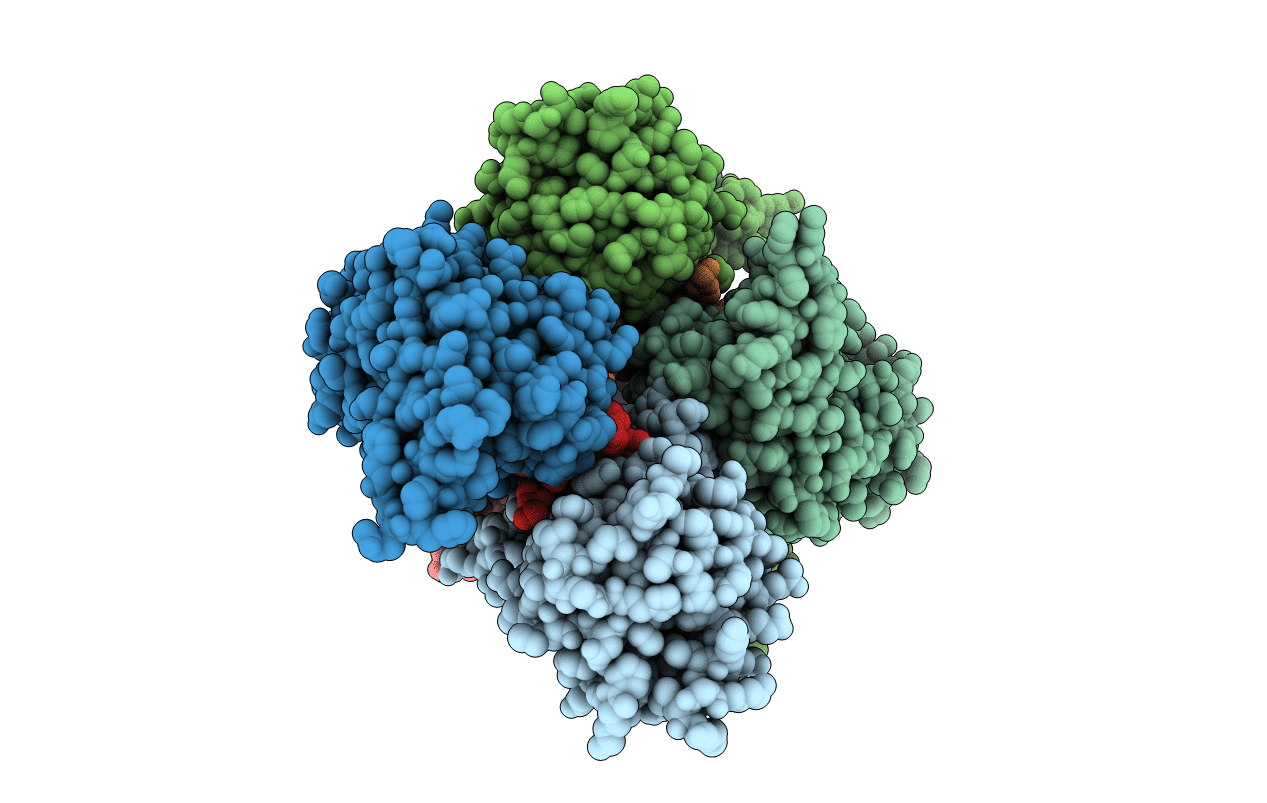 |
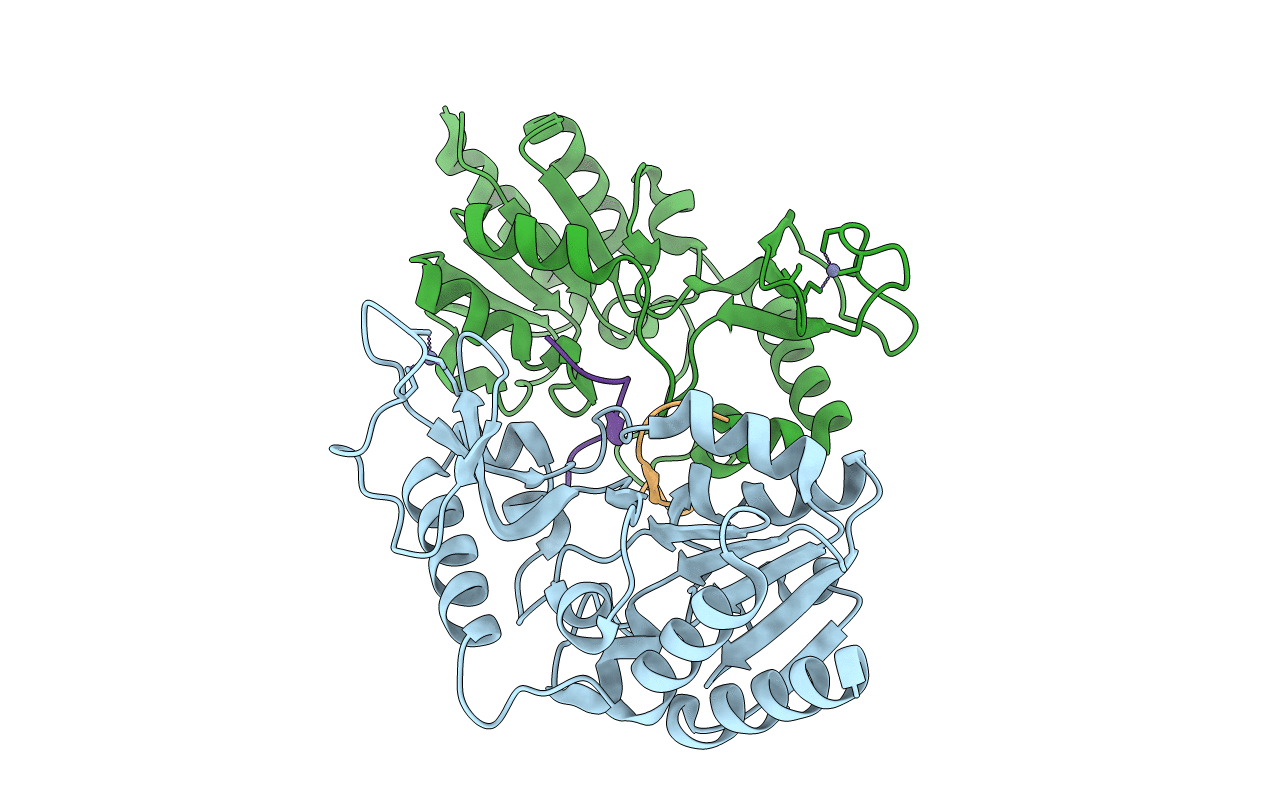
Crystal Structure Of Cobb Ac2 (A76G, I131C, V162G) In Complex With H4K16-Acetyl Peptide
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Cobb Ac2 (A76G, I131C, V162A) In Complex With H4K16-Buturyl Peptide
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: ZN |
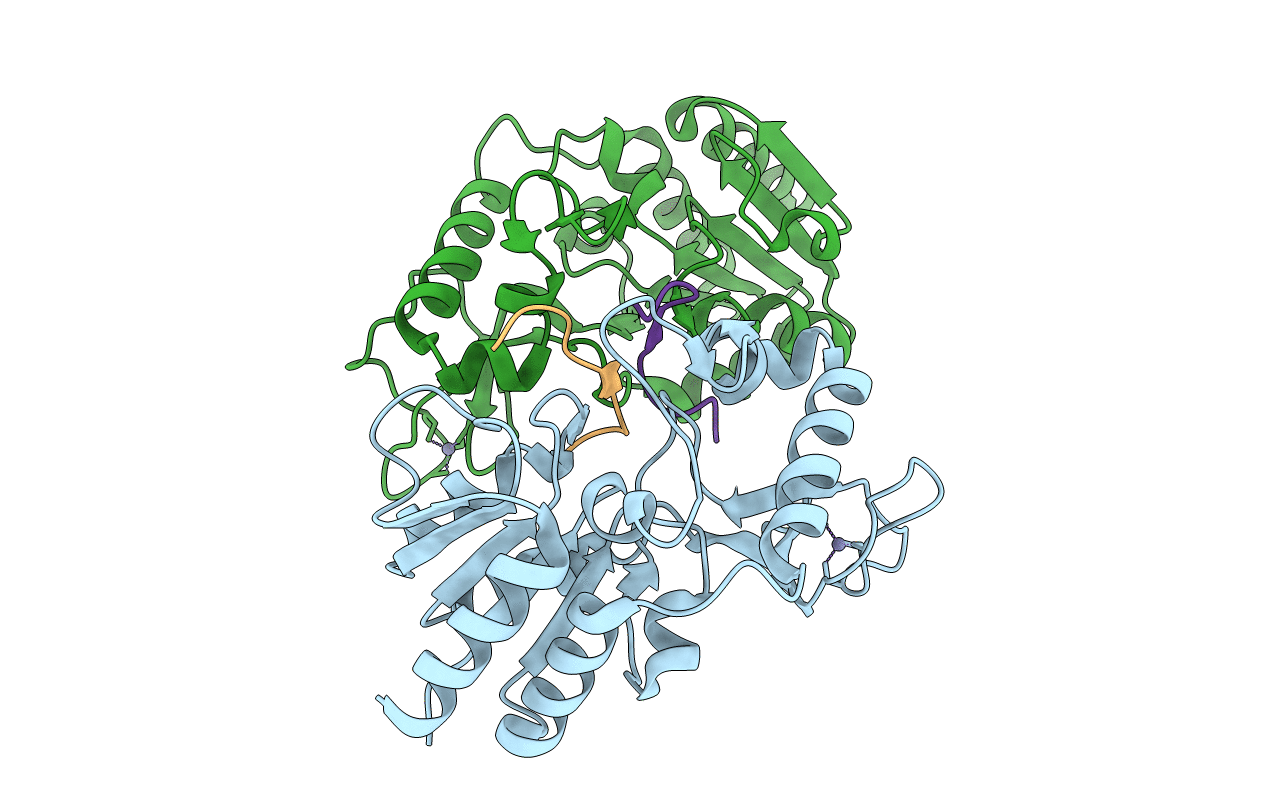 |
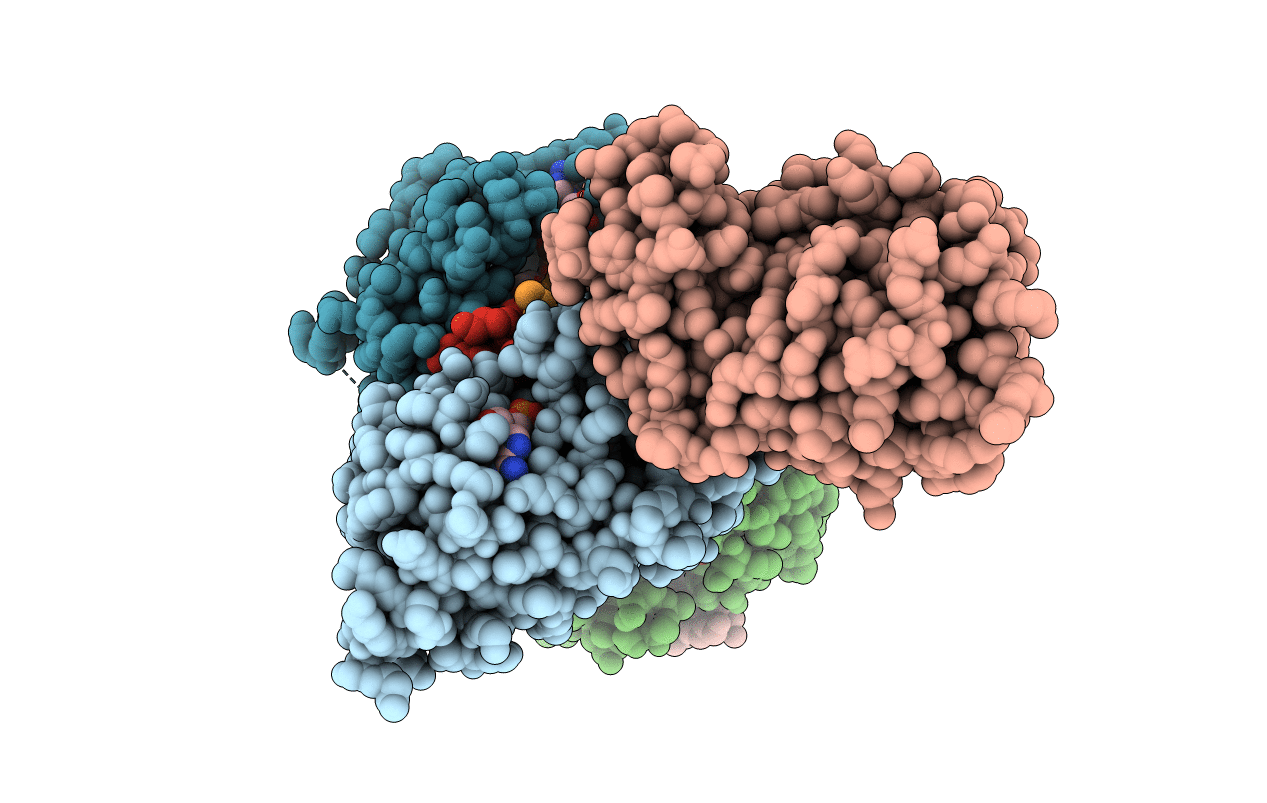
Crystal Structure Of Cobb Ac2 (A76G,I131C,V162A) In Complex With H4K16-Crotonyl Peptide
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: ZN |
 |
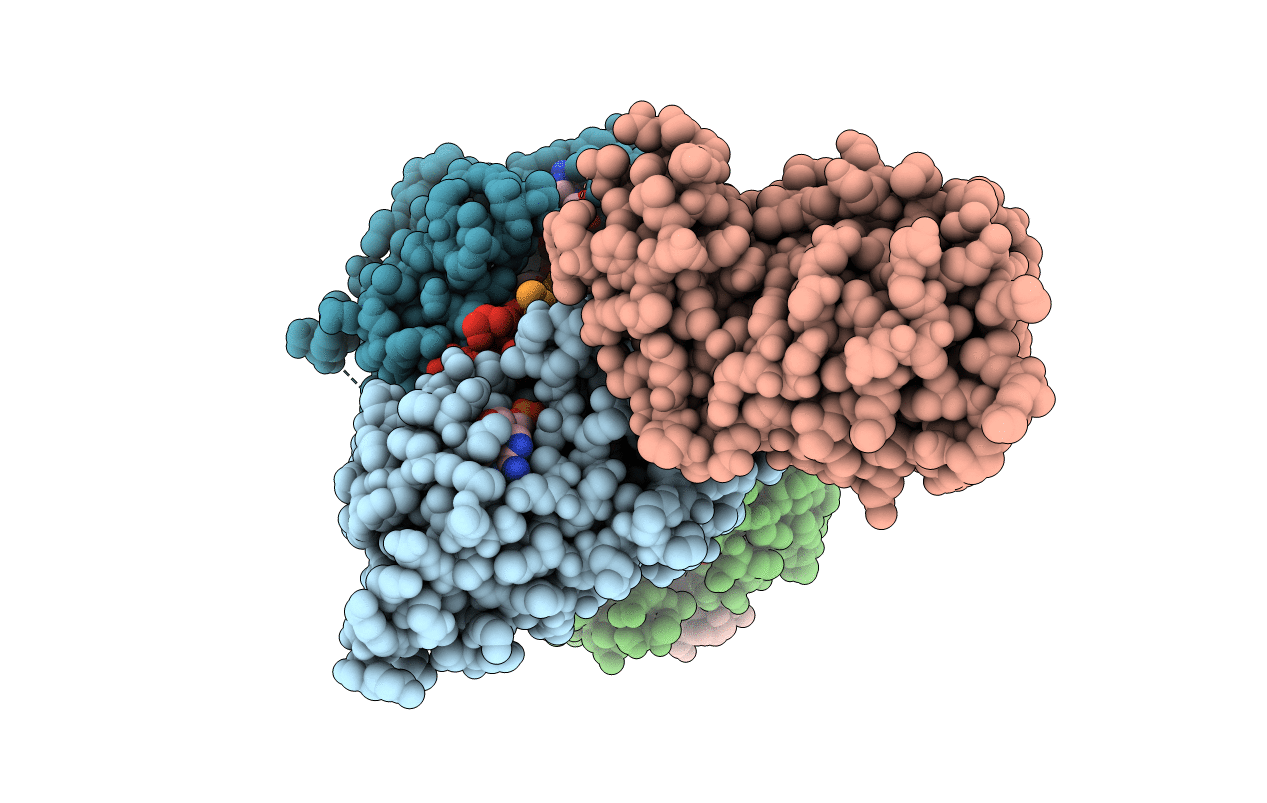
Crystal Structure Of Cobb Ac2 (A76G,I131C,V162A) In Complex With H4K16Cr-2'Oh-Adpr Peptide Intermediate After Soaking
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: KMQ |
 |
Crystal Structure Of Cobb Ac2 (A76G, I131C, V162G) In Complex With H4K16Cr-2'Oh-Adpr Peptide Intermediate After Co-Crystallisation
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: KMQ |
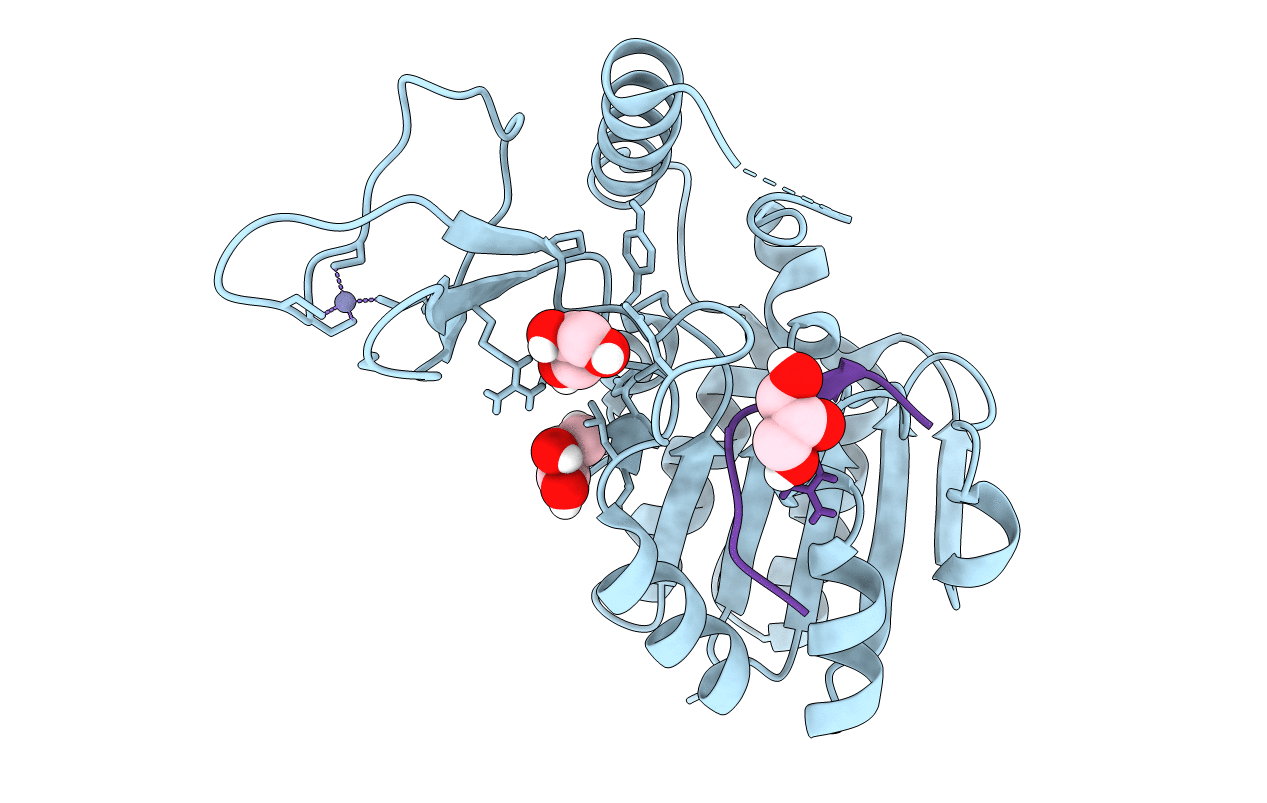 |
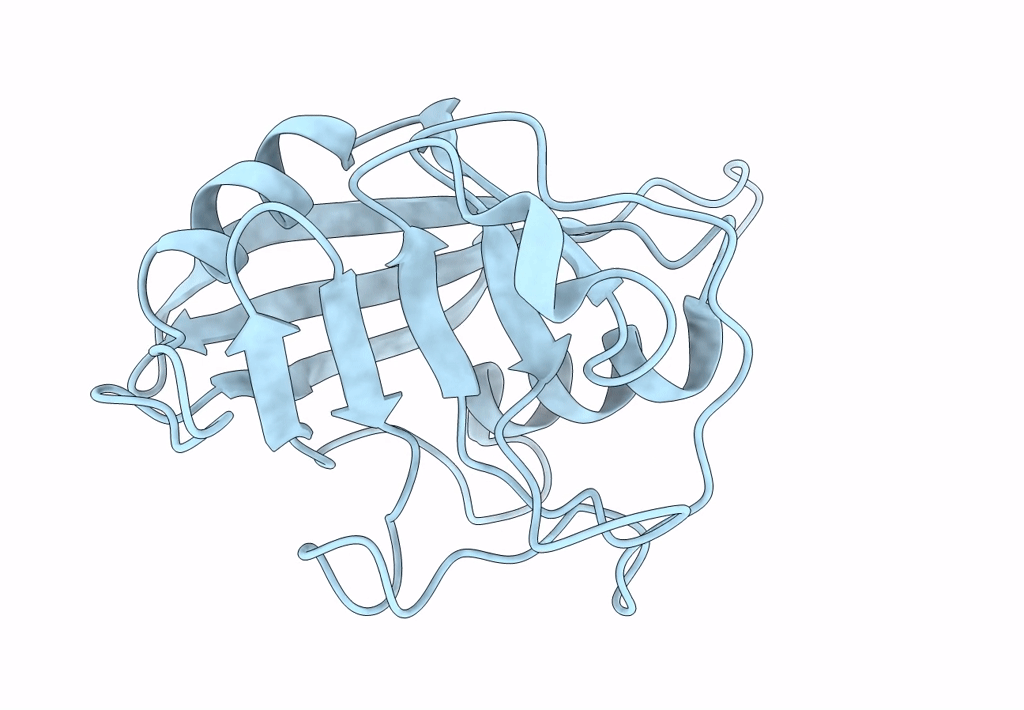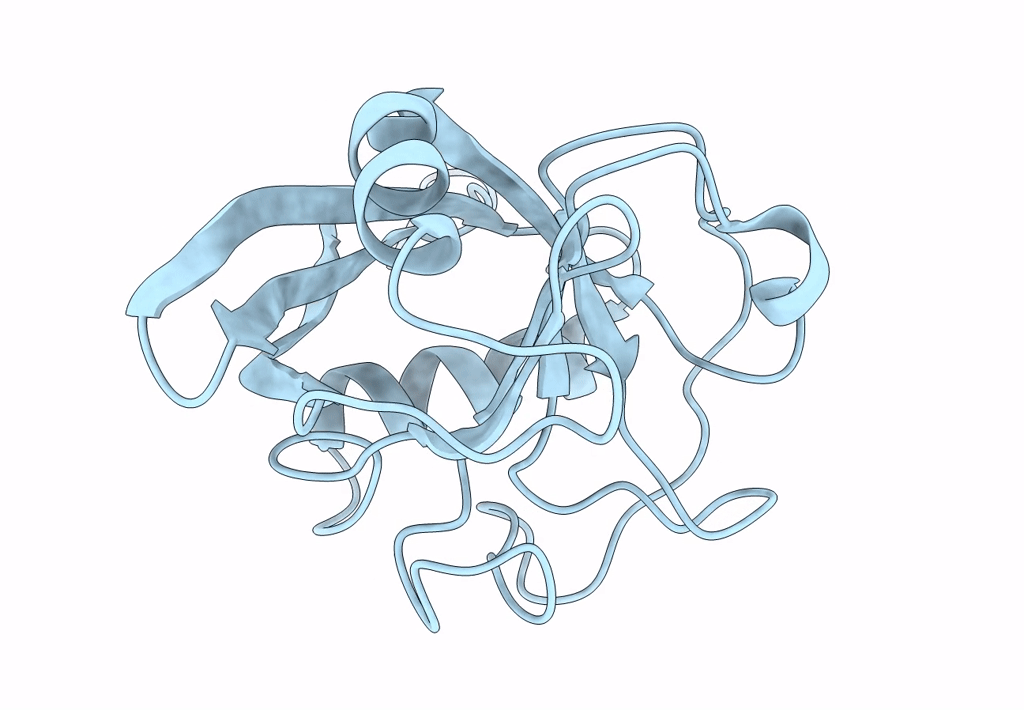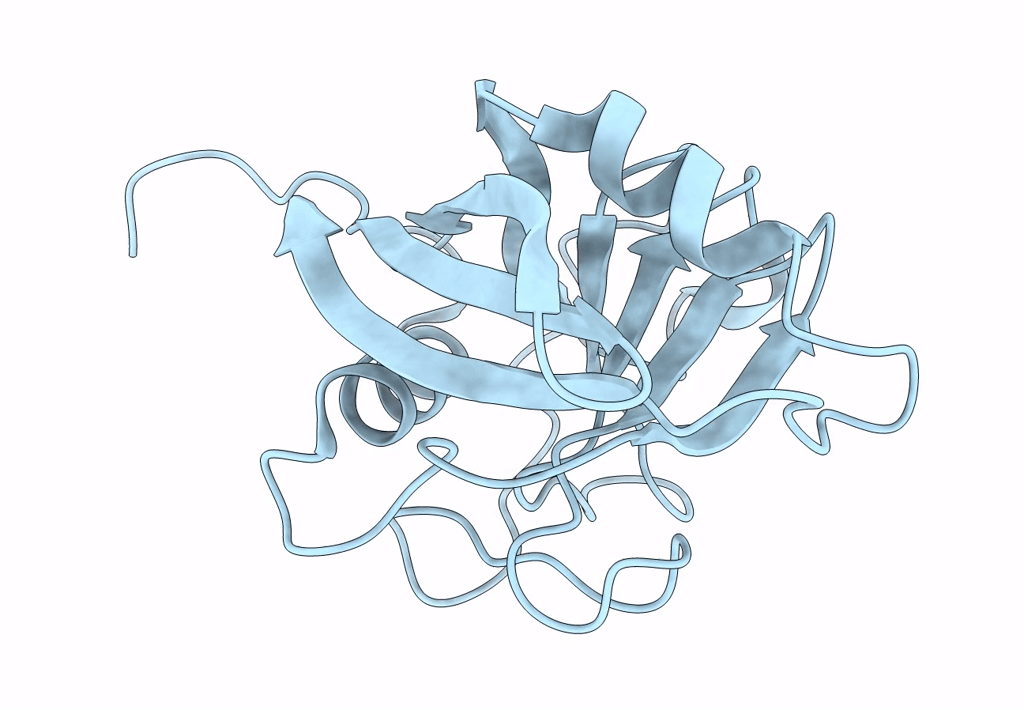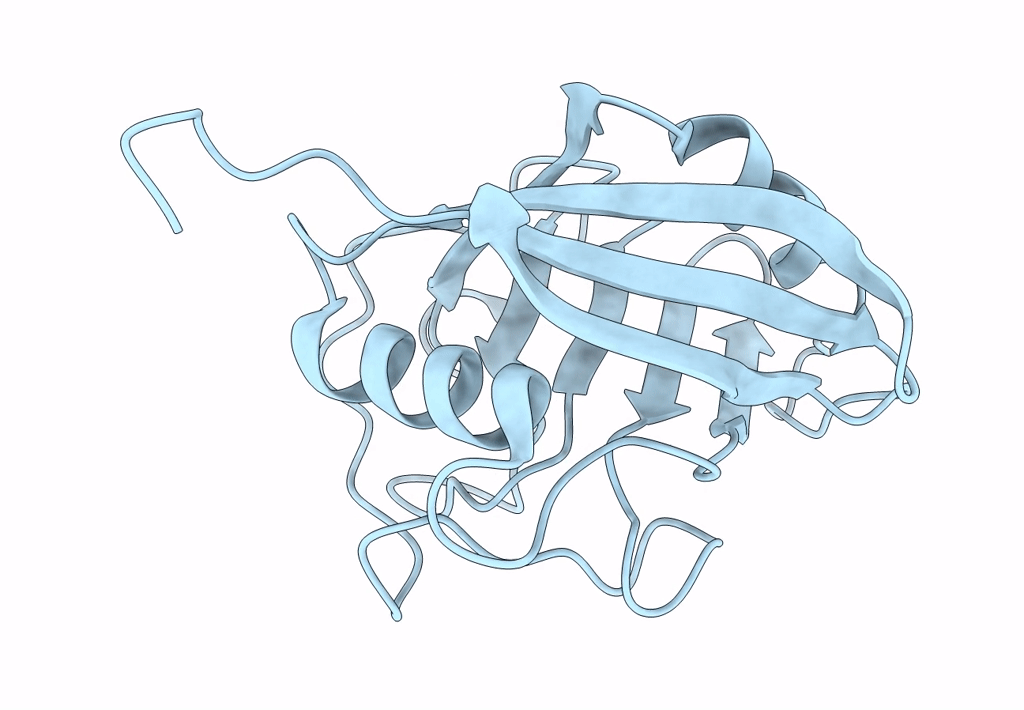
Crystal Structure Of Cobb Ac3(A76G,Y92A, I131L, V187Y) In Complex With H4K16-Acetyl Peptide
Organism: Escherichia coli (strain k12), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: ZN, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2010-03-23 Classification: ISOMERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-03-23 Classification: ISOMERASE Ligands: GOL, SO4 |
 |
Organism: Homo sapiens, Tolypocladium inflatum
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2010-03-23 Classification: ISOMERASE/IMMUNOSUPPRESSANT |
 |
Organism: Homo sapiens, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-03-23 Classification: ISOMERASE/VIRAL PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-05-05 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Crystal Structure Of A Eukaryotic Polyphosphate Polymerase In Complex With A Phosphate Polymer
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2009-05-05 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, SO4 |
 |
Crystal Structure Of A Eukaryotic Polyphosphate Polymerase In Complex With Appnhp-Mn2+
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-05-05 Classification: BIOSYNTHETIC PROTEIN Ligands: ANP, MN, SO4, NA |
 |
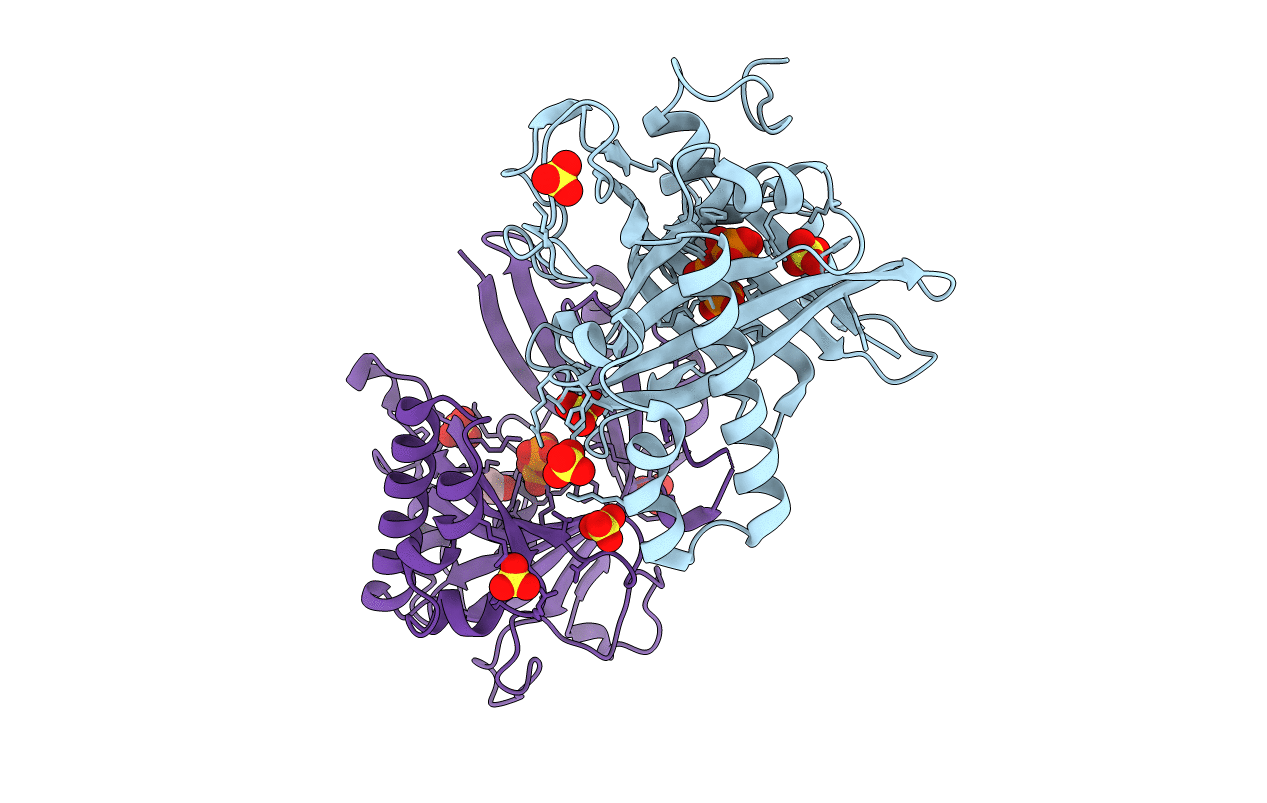
Crystal Structure Of A Eukaryotic Polyphosphate Polymerase In Complex With Orthophosphate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-05-05 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, EDO |
 |
Crystal Structure Of A Eukaryotic Polyphosphate Polymerase In Complex With Pyrophosphate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2009-05-05 Classification: BIOSYNTHETIC PROTEIN Ligands: POP, SO4, EDO |
 |
Solution Structure Of The Calmodulin Binding Domain (Cambd) Of Small Conductance Ca2+-Activated Potassium Channels (Sk2)
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 2001-12-14 Classification: SIGNALING PROTEIN |

