Search Count: 21
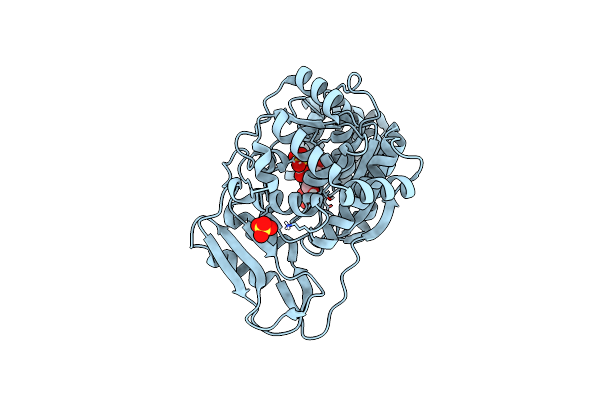 |
Crystal Structure Of Uridine Bound To Geobacillus Thermoglucosidasius Pyrimidine Nucleoside Phosphorylase Pynp
Organism: Parageobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-08-25 Classification: TRANSFERASE Ligands: SO4, URI |
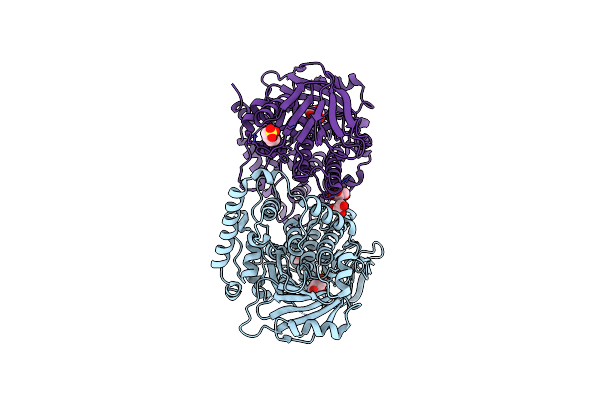 |
Organism: Brevundimonas sp. bal3
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-05-23 Classification: FLAVOPROTEIN Ligands: TLA, MES, GOL |
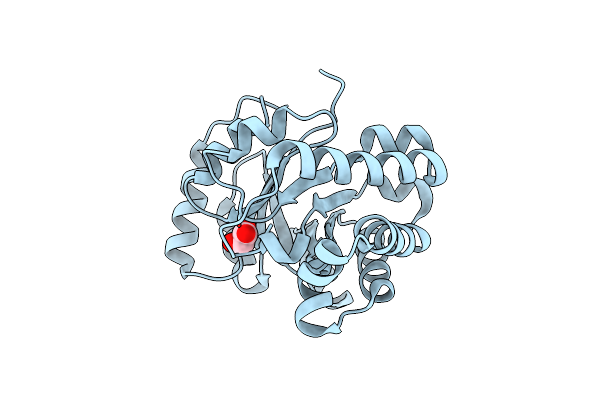 |
Structure-Based Protein Engineering Efforts On The Scaffold Of A Monomeric Triosephosphate Isomerase Yielding A Sugar Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-04-22 Classification: ISOMERASE Ligands: GOA |
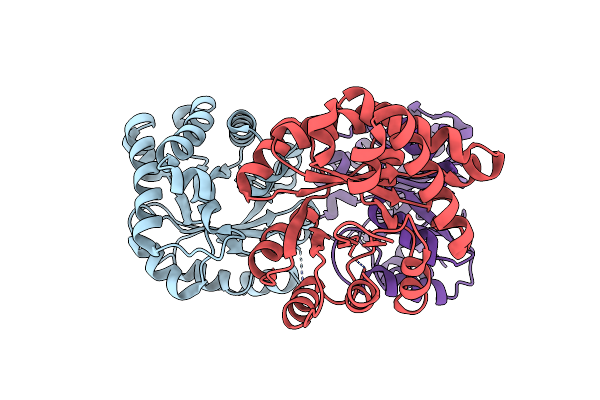 |
Structure-Based Protein Engineering Of A Monomeric Triosephosphate Isomerase Towards Changing Substrate Specificity
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2015-04-22 Classification: ISOMERASE |
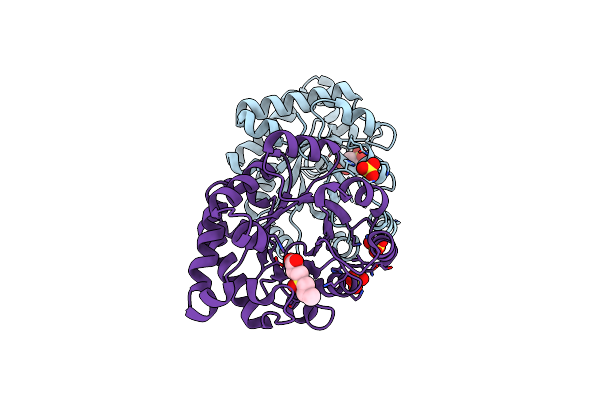 |
Crystallographic Binding Studies With An Engineered Monomeric Variant Of Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2010-01-26 Classification: ISOMERASE Ligands: SO4, X1R |
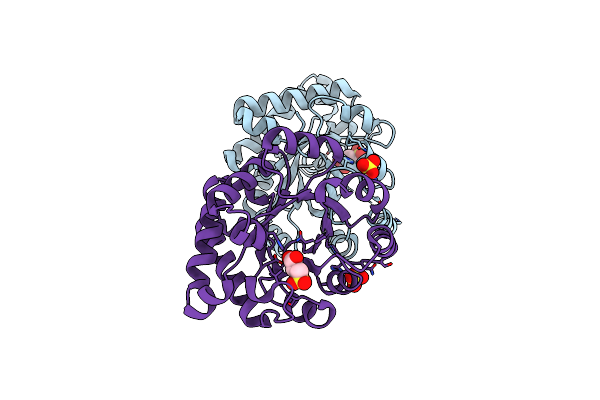 |
Crystallographic Binding Studies With An Engineered Monomeric Variant Of Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2010-01-26 Classification: ISOMERASE Ligands: X1S, SO4 |
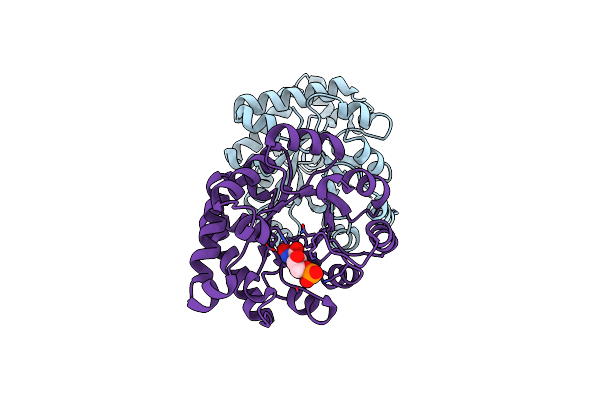 |
Crystallographic Binding Studies With An Engineered Monomeric Variant Of Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2010-01-26 Classification: ISOMERASE Ligands: RES |
 |
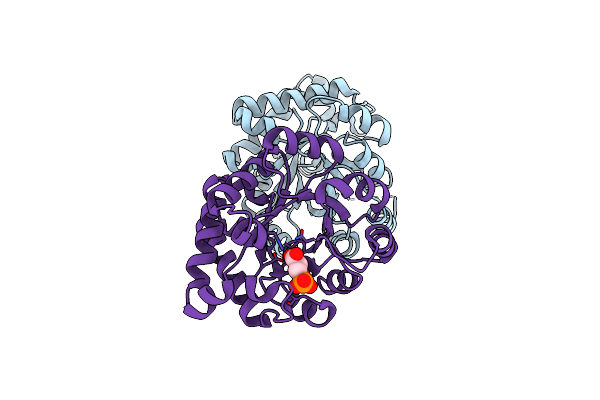
Crystallographic Binding Studies With An Engineered Monomeric Variant Of Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2010-01-26 Classification: ISOMERASE Ligands: SO4 |
 |
Crystallographic Binding Studies With An Engineered Monomeric Variant Of Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-01-26 Classification: ISOMERASE Ligands: 3PG |
 |
Crystallographic Binding Studies With An Engineered Monomeric Variant Of Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2009-12-29 Classification: ISOMERASE |
 |
The A178L Mutation In The C-Terminal Hinge Of The Flexible Loop-6 Of Triosephosphate Isomerase (Tim) Induces A More Closed Conformation Of This Hinge Region In Dimeric And Monomeric Tim
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-02-19 Classification: ISOMERASE Ligands: SO4, EPE |
 |
The A178L Mutation In The C-Terminal Hinge Of The Flexible Loop-6 Of Triosephosphate Isomerase (Tim) Induces A More Closed Conformation Of This Hinge Region In Dimeric And Monomeric Tim
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2008-02-19 Classification: ISOMERASE Ligands: SO4, PGA |
 |
The A178L Mutation In The C-Terminal Hinge Of The Flexible Loop-6 Of Triosephosphate Isomerase (Tim) Induces A More Closed Conformation Of This Hinge Region In Dimeric And Monomeric Tim
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-02-19 Classification: ISOMERASE Ligands: PO4 |
 |
The A178L Mutation In The C-Terminal Hinge Of The Flexible Loop-6 Of Triosephosphate Isomerase (Tim) Induces A More Closed Conformation Of This Hinge Region In Dimeric And Monomeric Tim
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2008-02-19 Classification: ISOMERASE Ligands: PGA, CL |
 |
Structure-Based Enzyme Engineering Efforts With An Inactive Monomeric Tim Variant: The Importance Of A Single Point Mutation For Generating An Active Site With Suitable Binding Properties
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2008-02-19 Classification: ISOMERASE Ligands: SO4 |
 |
Structure-Based Enzyme Engineering Efforts With An Inactive Monomeric Tim Variant: The Importance Of A Single Point Mutation For Generating An Active Site With Suitable Binding Properties
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-02-19 Classification: ISOMERASE Ligands: CIT, TBU, ASF |
 |
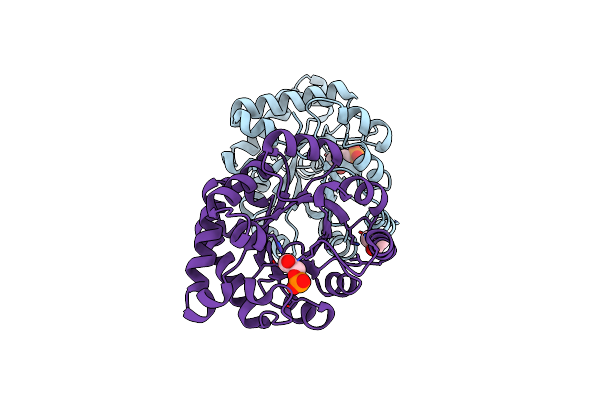
Structure-Based Enzyme Engineering Efforts With An Inactive Monomeric Tim Variant: The Importance Of A Single Point Mutation For Generating An Active Site With Suitable Binding Properties.
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-02-19 Classification: ISOMERASE Ligands: CL, PGA |
 |
Structure-Based Enzyme Engineering Efforts With An Inactive Monomeric Tim Variant: The Importance Of A Single Point Mutation For Generating An Active Site With Suitable Binding Properties
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-02-19 Classification: ISOMERASE Ligands: BBR, TBU |
 |
Structure-Based Enzyme Engineering Efforts With An Inactive Monomeric Tim Variant: The Importance Of A Single Point Mutation For Generating An Active Site With Suitable Binding Properties
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-02-19 Classification: ISOMERASE Ligands: CIT |
 |
The Functional Role Of The Conserved Active Site Proline Of Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-01-02 Classification: ISOMERASE |

