Search Count: 38
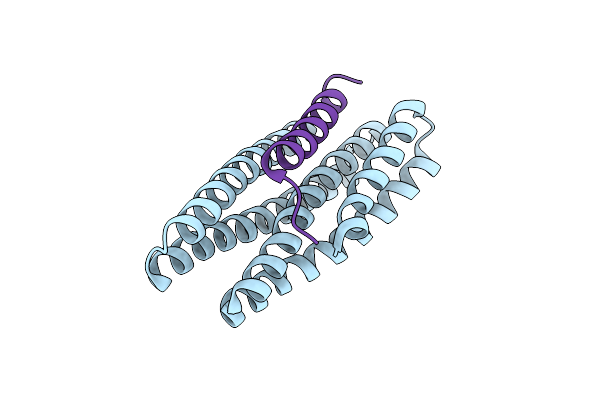 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
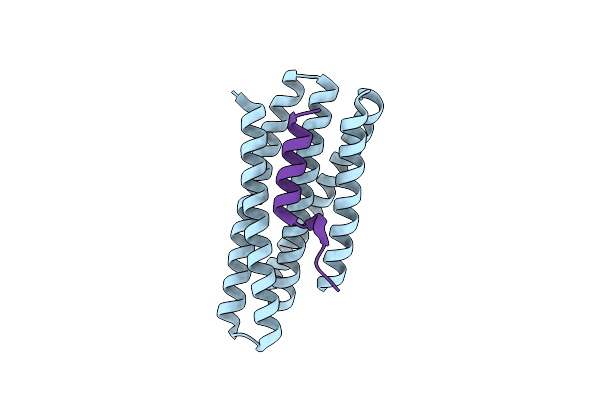 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
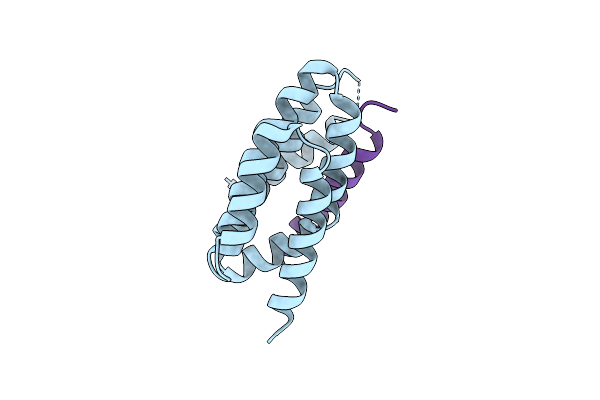 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
 |
De Novo Design Of High-Affinity Protein Binders To Bioactive Helical Peptides
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-01-10 Classification: DE NOVO PROTEIN |
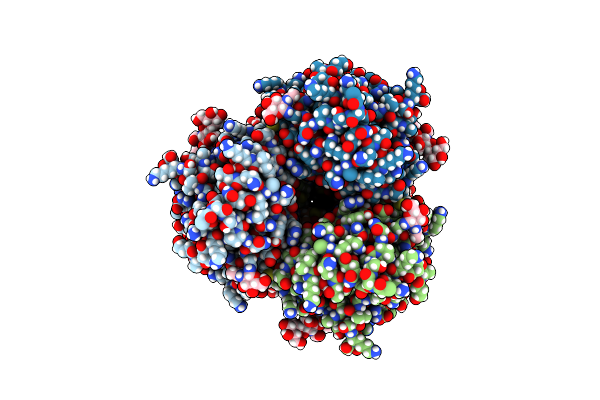 |
Cryo-Em Structure Of Designed Influenza Ha Binder, Ha_20, Bound To Influenza Ha (Strain: Iowa43)
Organism: Influenza a virus, Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: DE NOVO PROTEIN/Viral Protein Ligands: NAG |
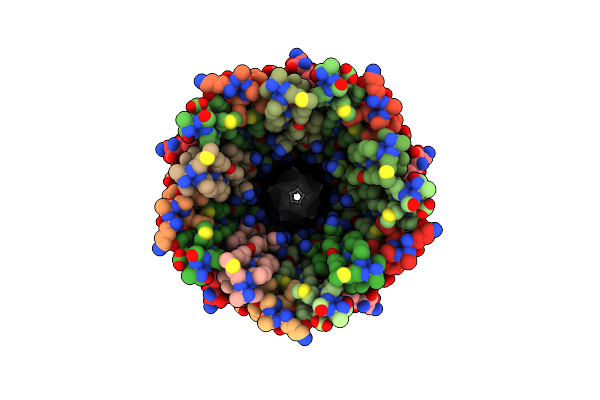 |
Cryoem Structure Of The Pointy Tip (Proteins Piii/Pvi/Pviii) From The F1 Filamentous Bacteriophage
Organism: Enterobacteria phage f1
Method: ELECTRON MICROSCOPY Resolution:2.97 Å Release Date: 2023-05-24 Classification: VIRUS |
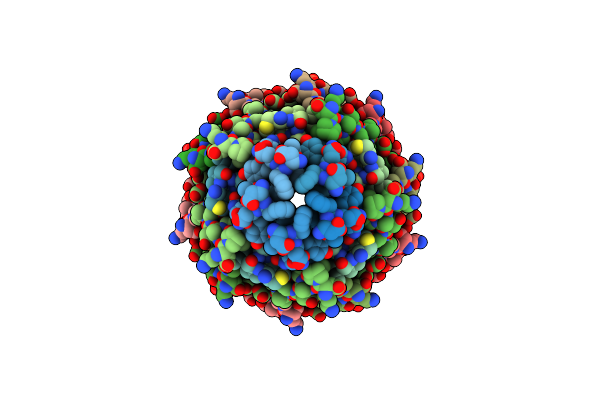 |
Cryoem Structure Of The Round Tip (Proteins Pvii/Pviii/Pix) From The F1 Filamentous Bacteriophage
Organism: Enterobacteria phage f1
Method: ELECTRON MICROSCOPY Resolution:2.81 Å Release Date: 2023-05-24 Classification: VIRUS |
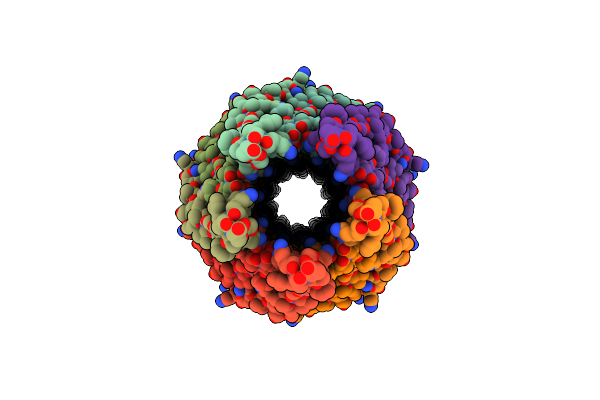 |
Cryoem Structure Of The Central Filamentous Region Of The F1 Filamentous Bacteriophage, Consisting Of The Major Capsid Protein Pviii
Organism: Enterobacteria phage f1
Method: ELECTRON MICROSCOPY Resolution:2.58 Å Release Date: 2023-05-24 Classification: VIRUS |
 |
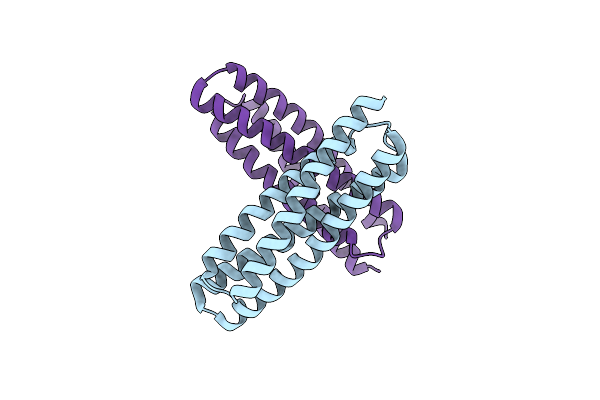
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-09-28 Classification: DE NOVO PROTEIN |
 |
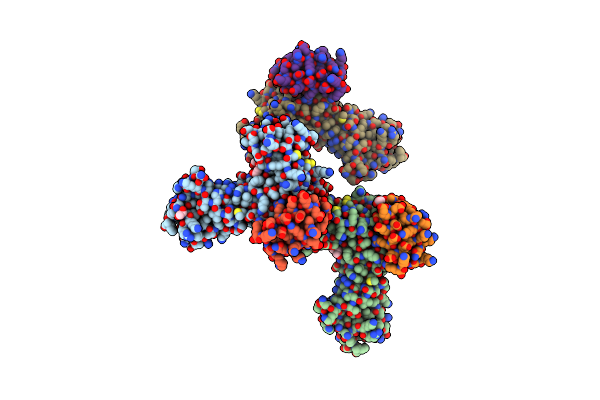
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2022-08-10 Classification: DE NOVO PROTEIN |
 |
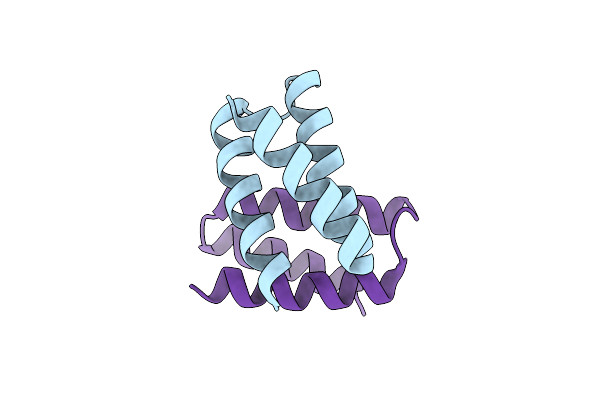
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-05-11 Classification: PROTEIN BINDING Ligands: PGE, EDO, PEG |
 |
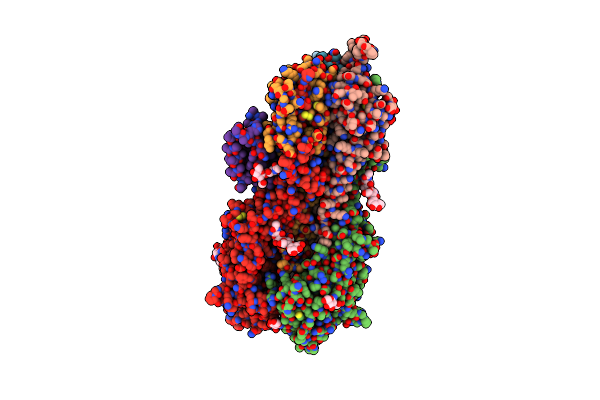
Unbound State Of A De Novo Designed Protein Binder To The Human Interleukin-7 Receptor
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-05-11 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of The De Novo Designed Binding Protein H3Mb In Complex With The 1968 Influenza A Virus Hemagglutinin
Organism: Influenza a virus (strain a/hong kong/1/1968 h3n2), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-05-04 Classification: VIRAL PROTEIN/DE NOVO PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-04-20 Classification: DE NOVO PROTEIN Ligands: NAG, EDO, SO4 |
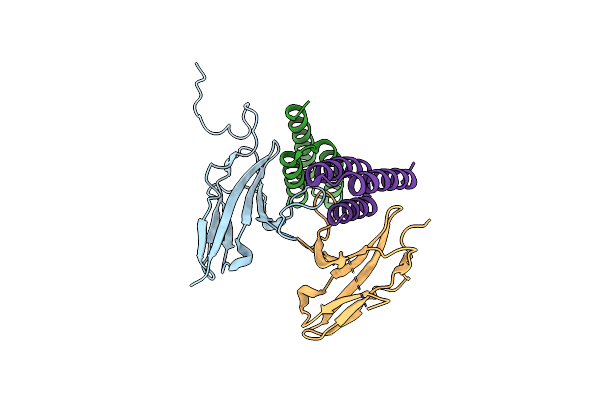 |
Crystal Structure Of Fgfr4 Domain 3 In Complex With A De Novo-Designed Mini-Binder
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-04-06 Classification: SIGNALING PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2022-04-06 Classification: DE NOVO PROTEIN |
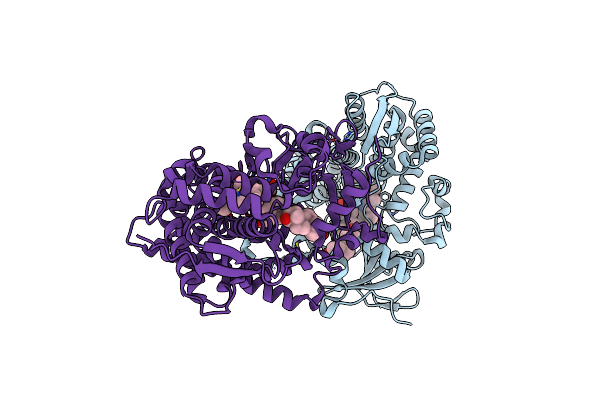 |
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2018-08-01 Classification: OXIDOREDUCTASE Ligands: HEM, ZN, TES, CL |
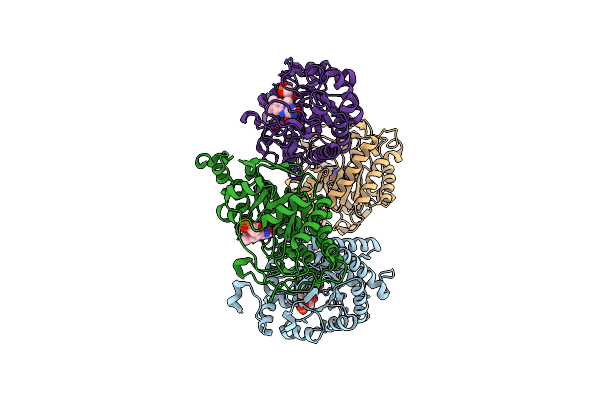 |
Organism: Thermus scotoductus sa-01
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-08-01 Classification: OXIDOREDUCTASE Ligands: FMN |
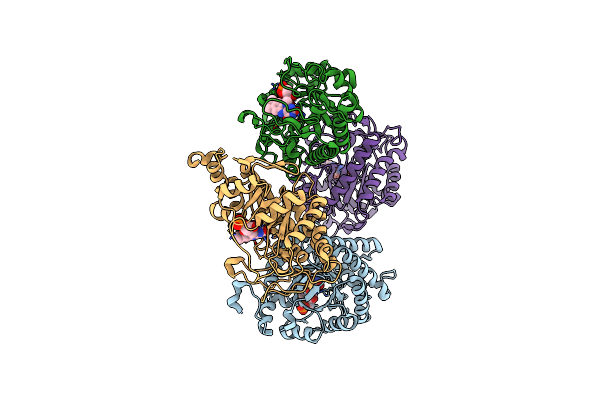 |
Organism: Thermus scotoductus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-05-30 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Crystal Structure Of Linalool/Nerolidol Synthase From Streptomyces Clavuligerus
Organism: Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2017-09-20 Classification: LIGASE Ligands: GOL |

