Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
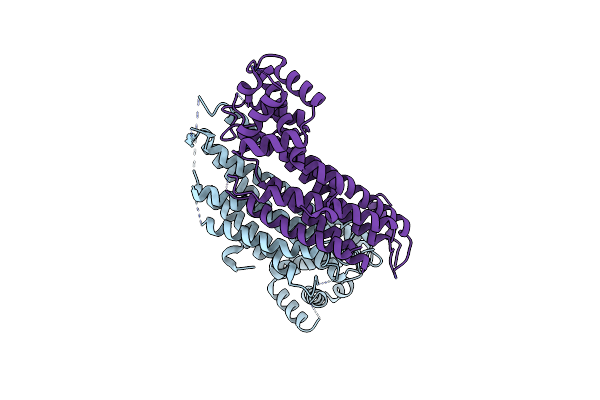 |
Crystal Structure Of Adenylosuccinate Lyase From Thermus Thermophilus Hb8, Ttpurb
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-11-15 Classification: LYASE |
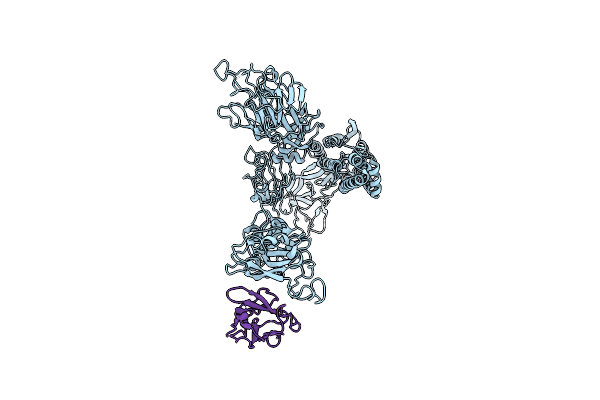 |
Minor Cryo-Em Structure Of S Protein Trimer Of Sars-Cov2 With K-874A Vhh, Composite Map
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: PROTEIN BINDING |
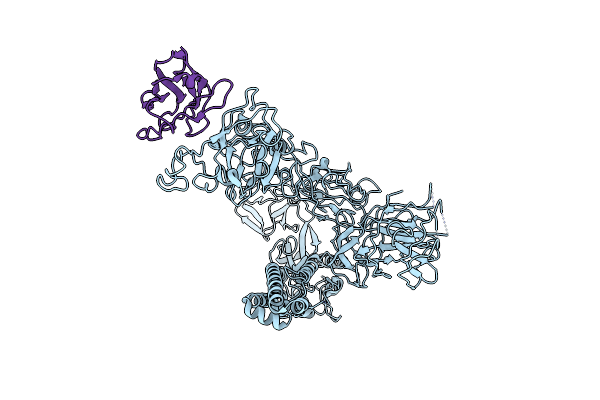 |
Major Cryo-Em Structure Of S Protein Trimer Of Sars-Cov2 With K-874, Composite Map
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: PROTEIN BINDING |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: PROTEIN BINDING |
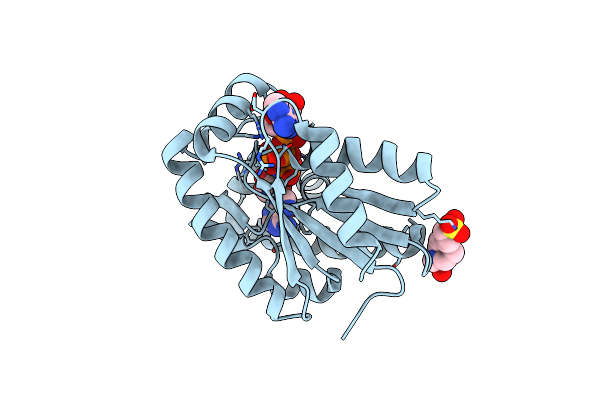 |
Crystal Structure Of Adenylate Kinase From An Extremophilic Archaeon Aeropyrum Pernix With Atp And Amp
Organism: Aeropyrum pernix (strain atcc 700893 / dsm 11879 / jcm 9820 / nbrc 100138 / k1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-22 Classification: TRANSFERASE Ligands: AMP, ATP, MG, EPE, CL |
 |
Crystal Structure Of (S)-3-O-Geranylgeranylglyceryl Phosphate Synthase From Thermoplasma Acidophilum In Complex With Substrate Sn-Glycerol-1-Phosphate
Organism: Thermoplasma acidophilum dsm 1728
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: 1GP |
 |
 |
 |
Crystal Structure Of Multiple Substrate Aminotransferase (Msat) From Thermococcus Profundus
Organism: Thermococcus profundus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2005-10-25 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2004-11-28 Classification: SIGNALING PROTEIN |
 |
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2004-11-28 Classification: Structural genomics, unknown function |
 |
Solution Structrue Of The Zf-An1 Domain From Arabidopsis Thaliana At2G36320 Protein
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2004-11-26 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Solution Structure Of The Zf-An1 Domain From Arabiopsis Thaliana F5O11.17 Protein
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2004-11-26 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Methanothermococcus thermolithotrophicus
Method: SOLUTION NMR Release Date: 2003-06-10 Classification: ISOMERASE |
 |
Organism: Halocynthia roretzi
Method: SOLUTION NMR Release Date: 2002-08-28 Classification: PROTEIN BINDING |
 |
 |
 |
Nmr Structure Of Intramolecular Dimer Antifreeze Protein Rd3, 40 Sa Structures
Organism: Pachycara brachycephalum
Method: SOLUTION NMR Release Date: 2001-02-28 Classification: ANTIFREEZE PROTEIN |
 |
Nmr Structure Of Intramolecular Dimer Antifreeze Protein Rd3, 40 Sa Structures
Organism: Pachycara brachycephalum
Method: SOLUTION NMR Release Date: 2001-02-28 Classification: ANTIFREEZE PROTEIN |