Search Count: 21
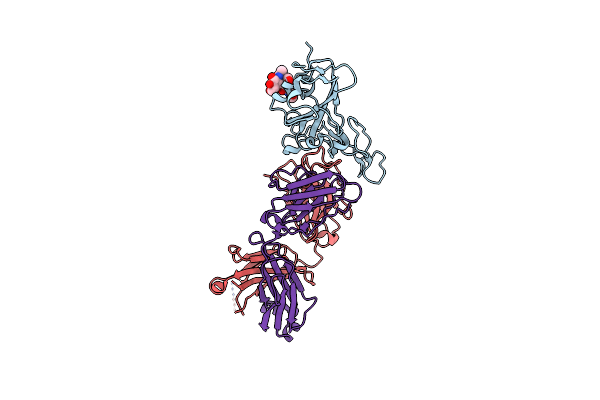 |
Crystal Structure Of Antibody Wrair-2123 In Complex With Sars-Cov-2 Receptor Binding Domain
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2024-04-17 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Pseudomonas monteilii
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-05-31 Classification: TOXIN Ligands: MG |
 |
Organism: Pseudomonas monteilii
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: TOXIN Ligands: MG |
 |
Organism: Pseudomonas monteilii
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: TOXIN Ligands: MG |
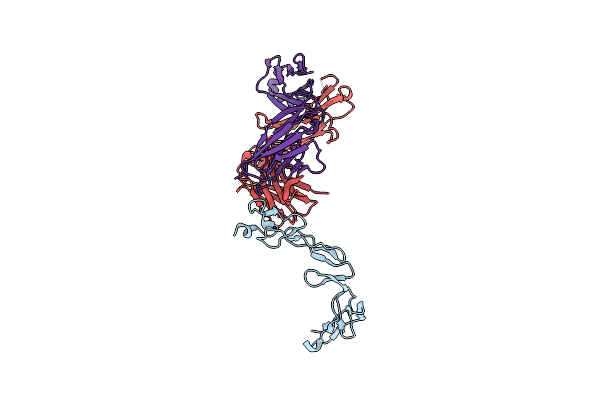 |
Crystal Structure Of Agonistic Antibody 1618 Fab Domain Bound To Human 4-1Bb.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2023-01-25 Classification: IMMUNE SYSTEM Ligands: TMO, MN |
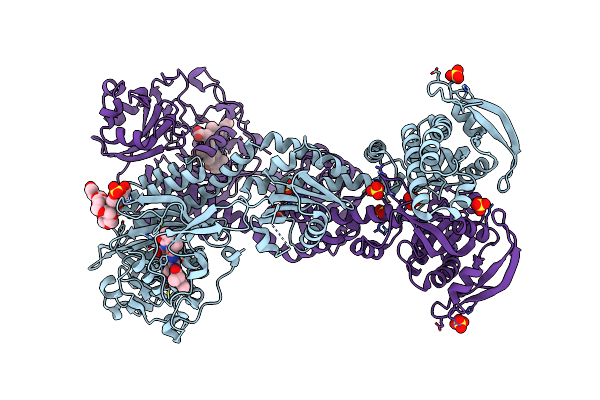 |
Crystal Structure Of An Artificial Phytochrome Regulated Adenylate/Guanylate Cyclase In Its Dark Adapted Pr Form
Organism: Deinococcus radiodurans, Synechocystis sp. pcc 6803 substr. kazusa
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-04-18 Classification: LYASE Ligands: LBV, SO4, 12P |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-06-14 Classification: TOXIN |
 |
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-10-09 Classification: CELL ADHESION |
 |
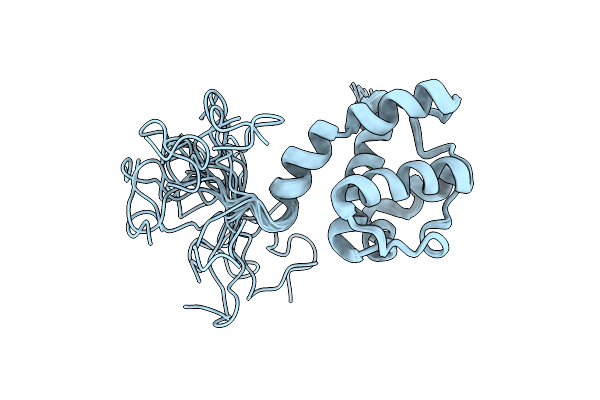
Ras Signaling Requires Dynamic Properties Of Ets1 For Phosphorylation-Enhanced Binding To Co-Activator Cbp
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2010-05-05 Classification: TRANSCRIPTION, PROTEIN BINDING |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2008-04-15 Classification: HYDROLASE Ligands: PO4, DK2 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-04-15 Classification: HYDROLASE Ligands: DMS, OX6, SO4, OX7 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-04-15 Classification: HYDROLASE Ligands: PO4, 23C |
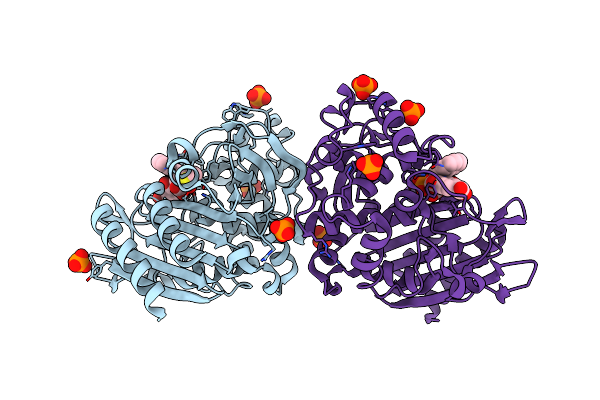 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-04-15 Classification: HYDROLASE Ligands: PO4, WH6, DMS |
 |
 |
 |
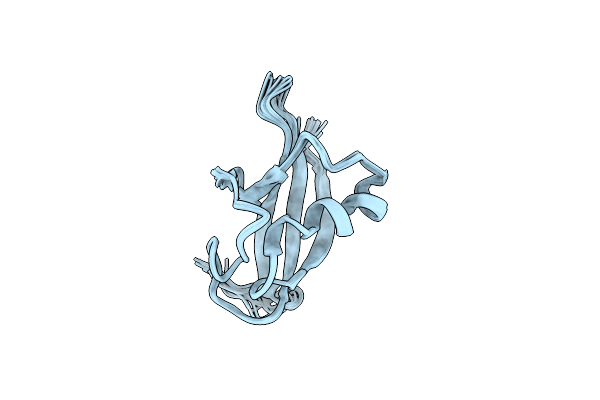
Nmr Structure Of The Nonstructural Protein 7 (Nsp7) From The Sars Coronavirus
Organism: Sars coronavirus
Method: SOLUTION NMR Release Date: 2005-12-06 Classification: VIRAL PROTEIN |
 |
X-Ray Structure Of Calcium-Loaded Calbindomodulin (A Calbindin D9K Re-Engineered To Undergo A Conformational Opening) At 1.44 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2004-05-25 Classification: SIGNALING PROTEIN Ligands: CA, ZN |
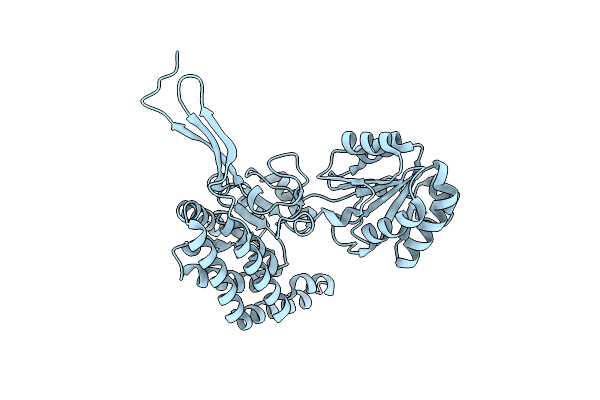 |
Crystal Structure Of Gamma-Glutamyl Phosphate Reductase (Tm0293) From Thermotoga Maritima At 2.00 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-04-01 Classification: OXIDOREDUCTASE |
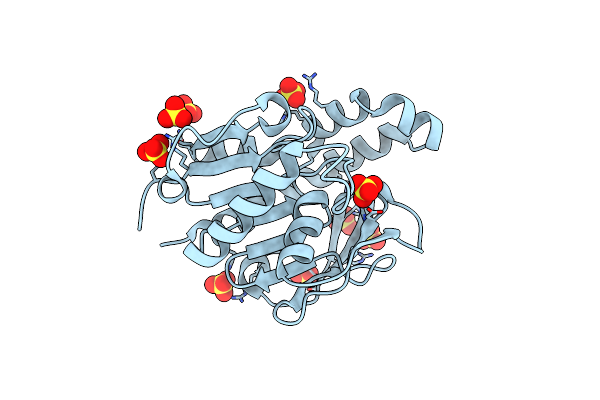 |
Crystal Structure Of A Glutamine Amidotransferase (Tm1158) From Thermotoga Maritima At 1.70 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2003-03-11 Classification: TRANSFERASE Ligands: SO4 |
 |
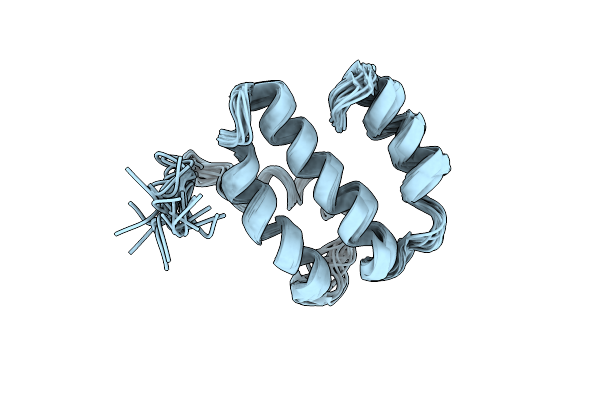
Organism: Bos taurus
Method: SOLUTION NMR Release Date: 2001-11-21 Classification: METAL BINDING PROTEIN |

