Search Count: 18
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2016-12-14 Classification: DE NOVO PROTEIN Ligands: PO4, GOL |
 |
Organism: Synthetic construct, Macaca fascicularis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-12-14 Classification: CYTOKINE/DE NOVO PROTEIN Ligands: ACT |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-11-02 Classification: DE NOVO PROTEIN Ligands: GOL |
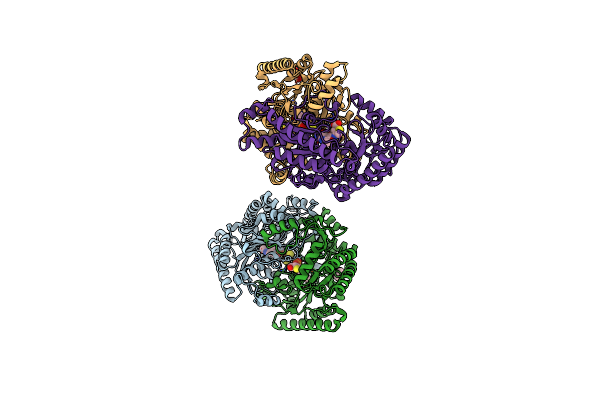 |
Gaba-Aminotransferase Inactivated By Conformationally-Restricted Inactivator
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: PSZ, FES, GOL |
 |
Fusion To A Highly Stable Consensus Albumin Binding Domain Allows For Tunable Pharmacokinetics
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2015-09-02 Classification: DE NOVO PROTEIN |
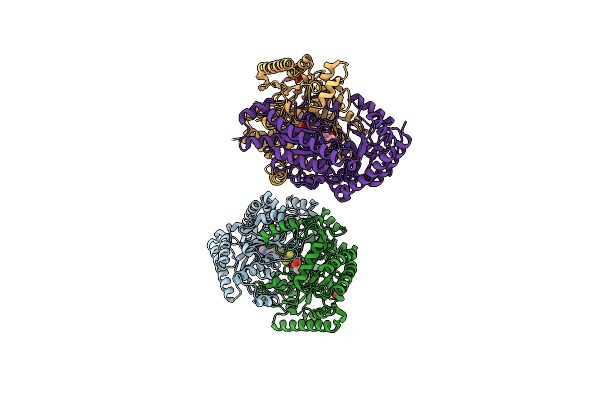 |
Gamma-Aminobutyric Acid Aminotransferase Inactivated By (1S,3S)-3-Amino-4-Difluoromethylenyl-1-Cyclopentanoic Acid (Cpp-115)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2015-03-11 Classification: TRANSFERASE Ligands: FES, PLP, GOL |
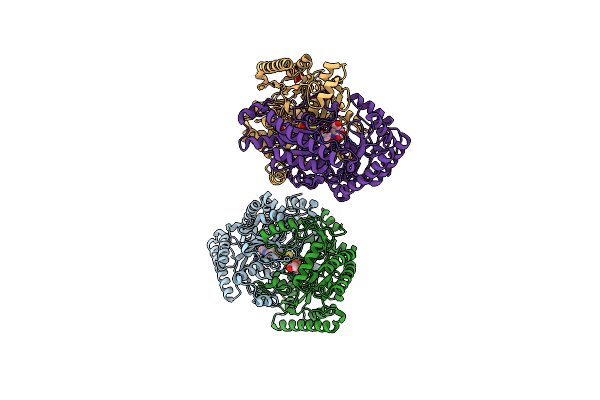 |
Gamma-Aminobutyric Acid Aminotransferase Inactivated By (1S,3S)-3-Amino-4-Difluoromethylenyl-1-Cyclopentanoic Acid (Cpp-115)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2015-02-25 Classification: TRANSFERASE Ligands: FES, RW2, GOL |
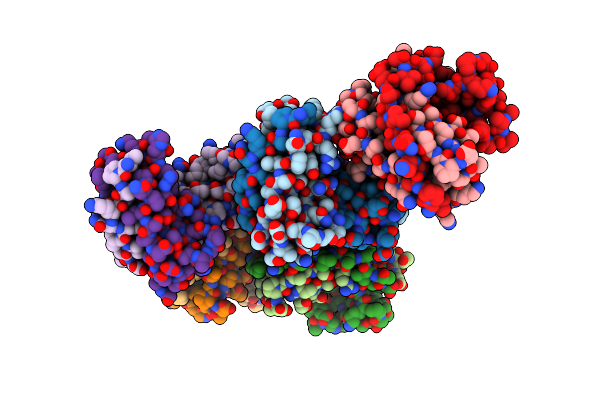 |
N-Terminal Beta-Strand Swapping In A Consensus Derived Alternative Scaffold Driven By Stabilizing Hydrophobic Interactions
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2014-02-26 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2014-01-29 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2014-01-29 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-01-29 Classification: DE NOVO PROTEIN Ligands: TRS |
 |
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-01-29 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-01-29 Classification: DE NOVO PROTEIN |
 |
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2014-01-29 Classification: DE NOVO PROTEIN |
 |
Method: X-RAY DIFFRACTION
Resolution:2.50 Å Release Date: 2012-05-16 Classification: DE NOVO PROTEIN |
 |
Method: X-RAY DIFFRACTION
Resolution:1.00 Å Release Date: 2012-05-16 Classification: DE NOVO PROTEIN Ligands: DIO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-04-28 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2009-04-07 Classification: IMMUNE SYSTEM Ligands: SO4 |

