Search Count: 35
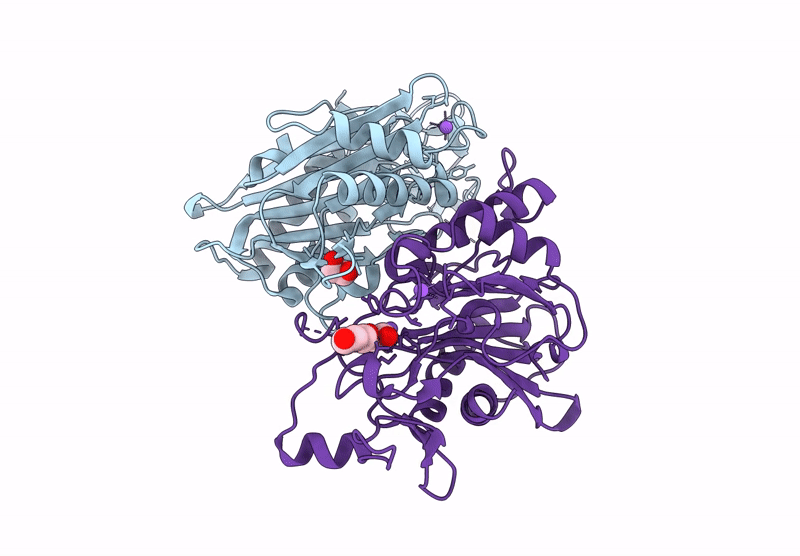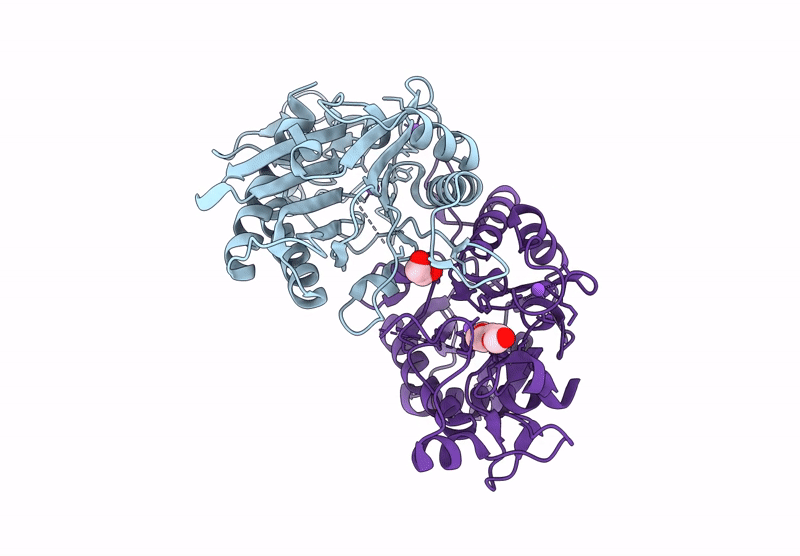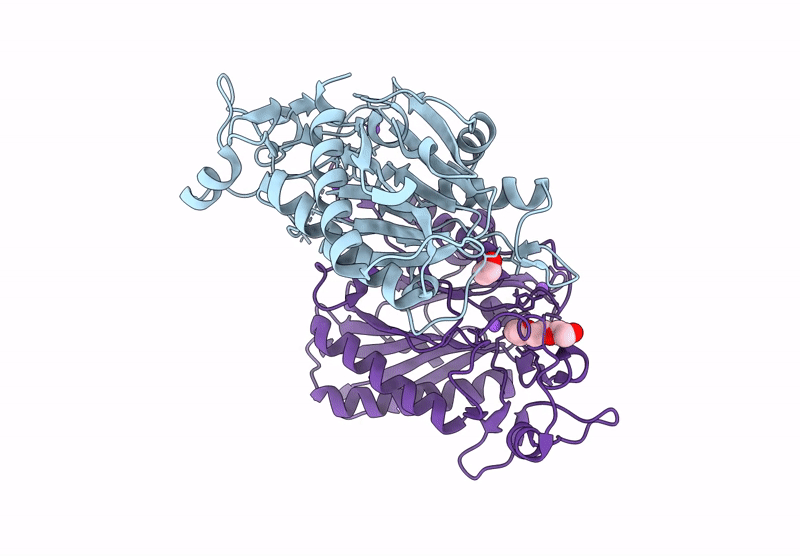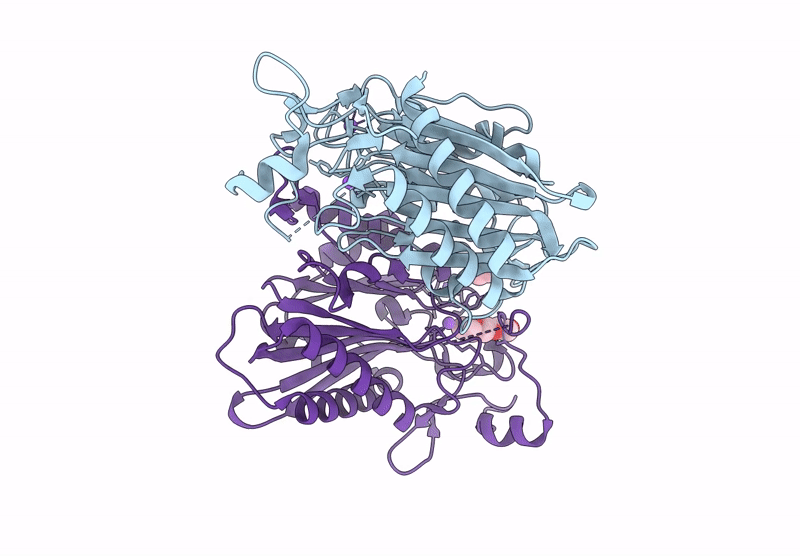 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PGE |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature Bound To Tetraethylene Glycol
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PG4 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase Nylc, D122G/H130Y/T267C Mutant, Hydroxylamine-Treated
Organism: Arthrobacter
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2023-03-29 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase Nylc Precursor, H130Y/N266A/T267A Mutant
Organism: Arthrobacter
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-03-01 Classification: HYDROLASE Ligands: GOL, SO4, NA |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase Nylc Precursor, D122G/H130Y/T267C Mutant
Organism: Arthrobacter
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2023-03-01 Classification: HYDROLASE Ligands: GOL, NA, SO4 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72.
Organism: Arthrobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: GOL, SO4, CL |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D122G Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: SO4, CL, GOL |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D122R Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D122K Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D122V Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: GOL, PO4 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., H130Y Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D122G/H130Y Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2018-07-25 Classification: HYDROLASE Ligands: GOL, NA, SO4 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase From Arthrobacter Sp. Ki72., D36A/D122G/H130Y/E263Q Mutant
Organism: Flavobacterium sp. ki723t1
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2018-07-25 Classification: HYDROLASE Ligands: GOL, PO4, CL |
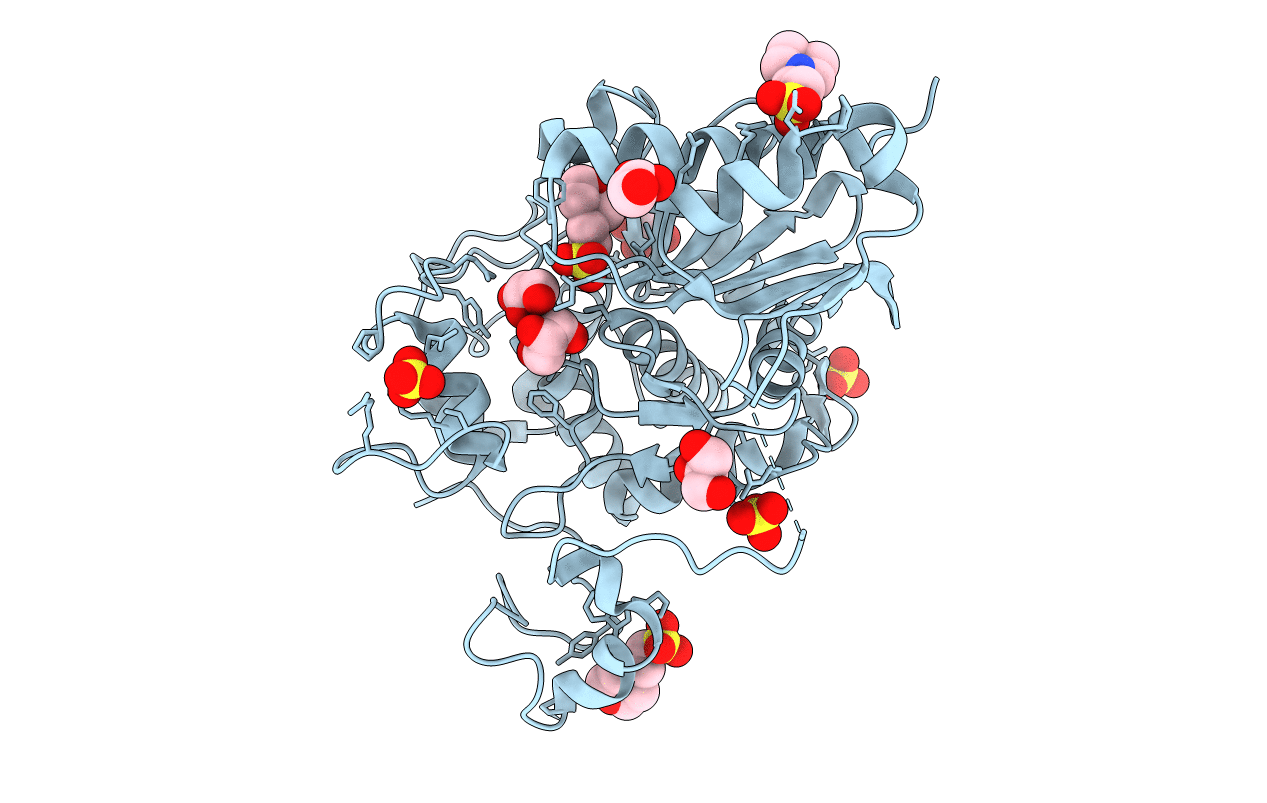 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase G181D/R187S/H266N/D370Y Mutant
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: GOL, MES, SO4 |
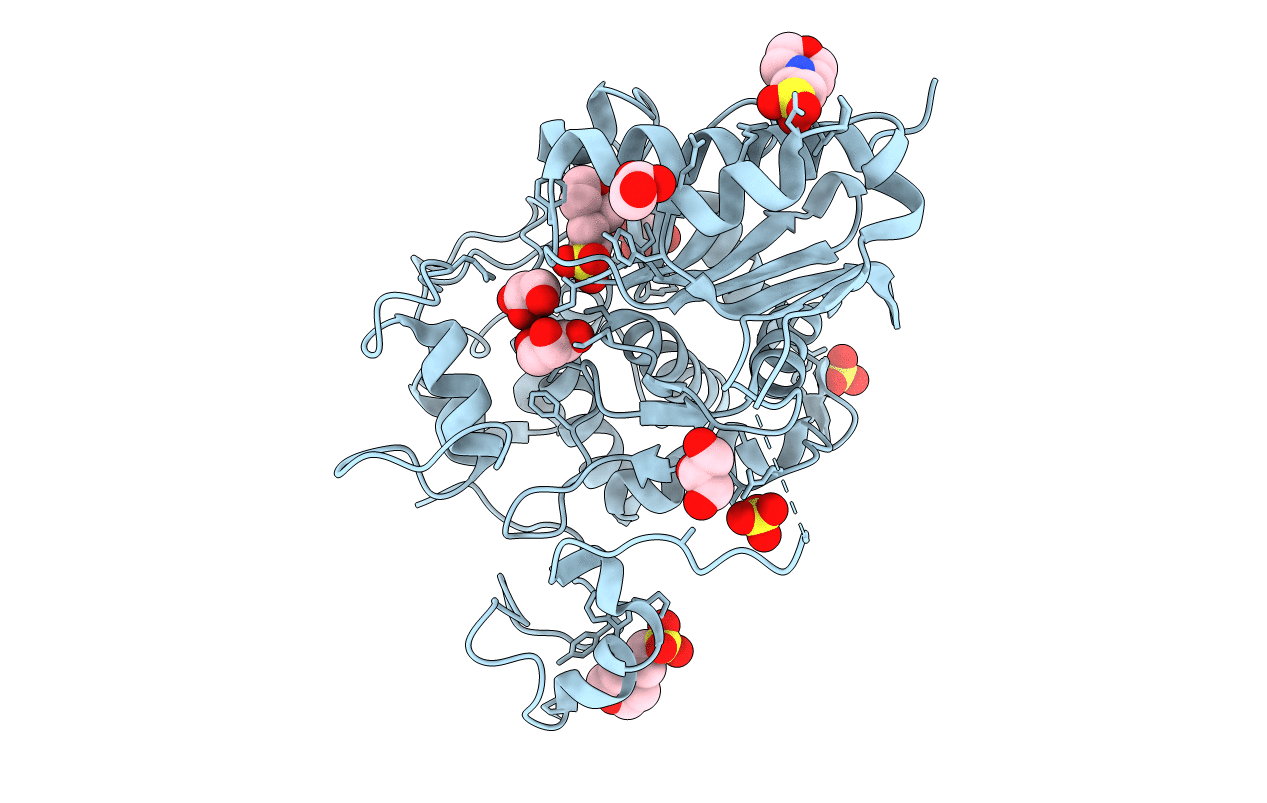 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase G181D/R187A/H266N/D370Y Mutant
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: GOL, MES, SO4 |
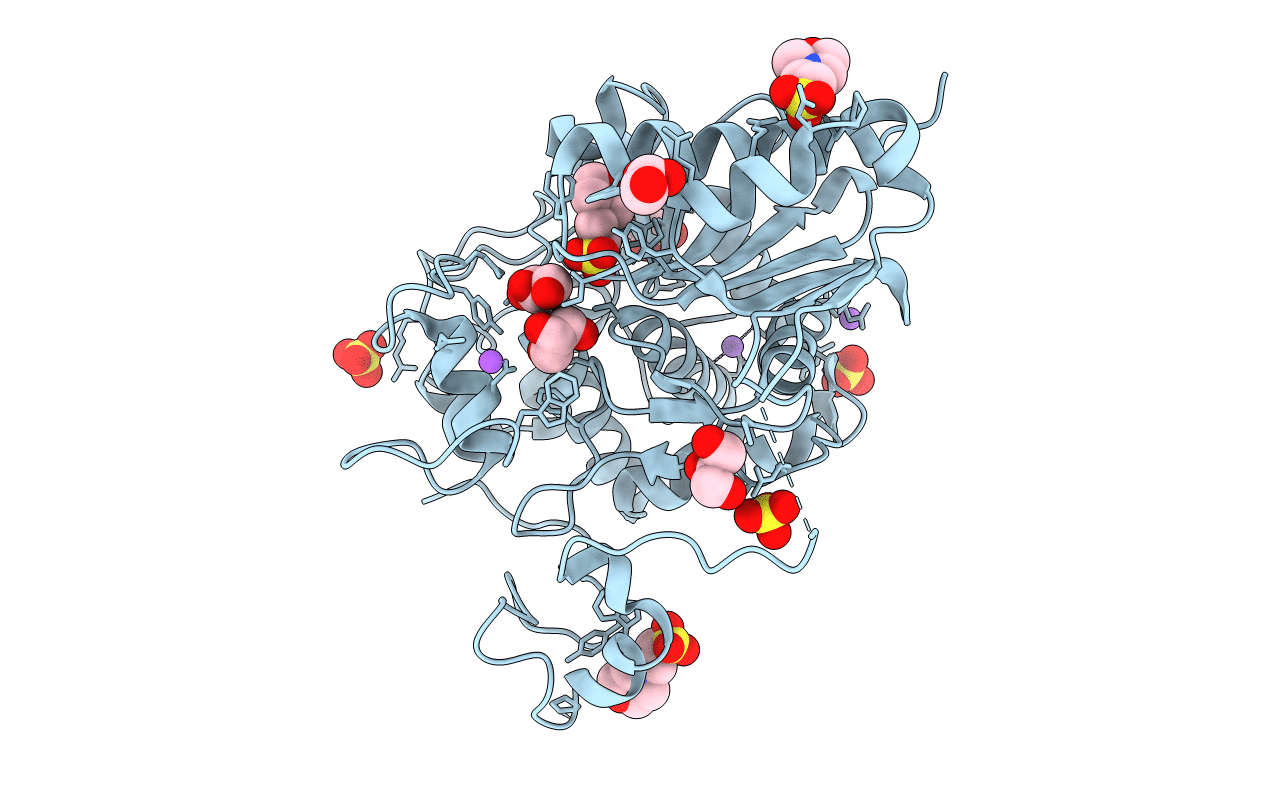 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase G181D/R187G/H266N/D370Y Mutant
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: GOL, MES, SO4, NA |
 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/R187S/H266N/D370Y Mutant Complexd With 6-Aminohexanoate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-10-16 Classification: HYDROLASE |
 |
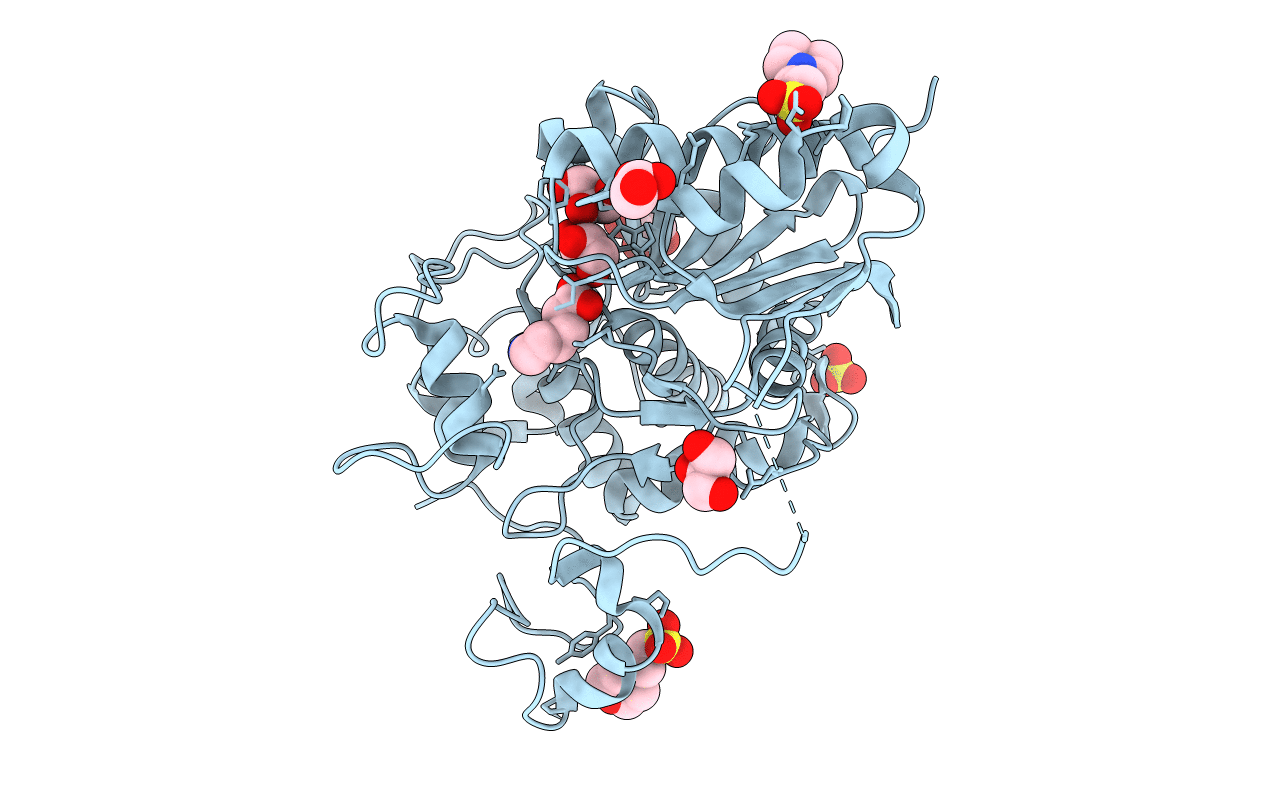
6-Aminohexanoate-Dimer Hydrolase S112A/G181D/R187A/H266N/D370Y Mutant Complexd With 6-Aminohexanoate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: ACA, GOL, MES |
 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/R187G/H266N/D370Y Mutant Complexd With 6-Aminohexanoate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: ACA, GOL, MES, SO4 |
 |
Organism: Agromyces
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-12-21 Classification: HYDROLASE |

