Search Count: 8
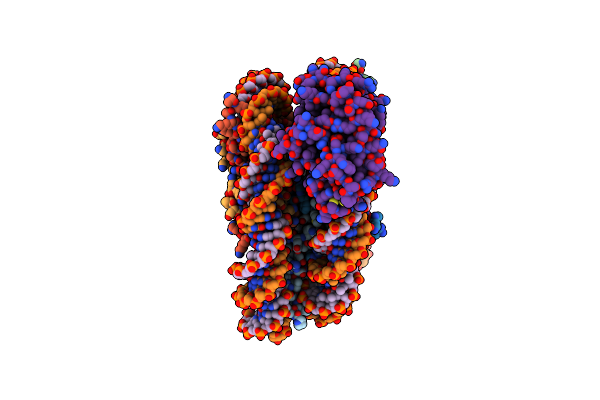 |
Organism: Saccharomyces cerevisiae, Xenopus laevis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
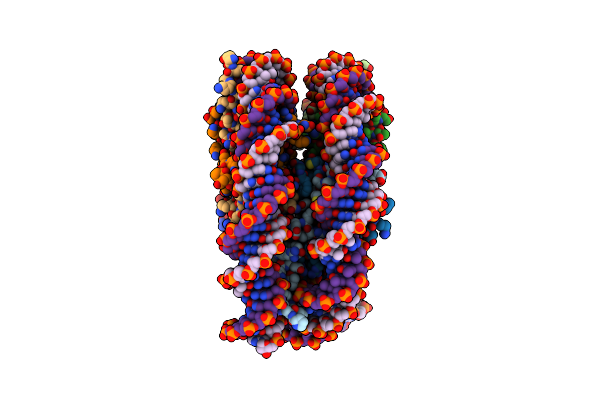 |
Organism: Xenopus laevis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: DNA BINDING PROTEIN/DNA |
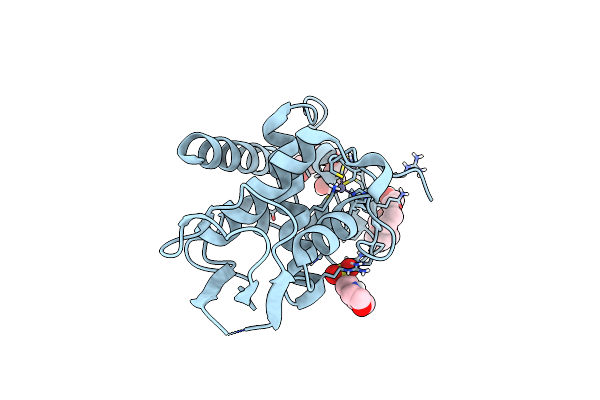 |
Organism: Vanderwaltozyma polyspora dsm 70294
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-03-29 Classification: DNA BINDING PROTEIN Ligands: ZN, MES, 1PE, PG4 |
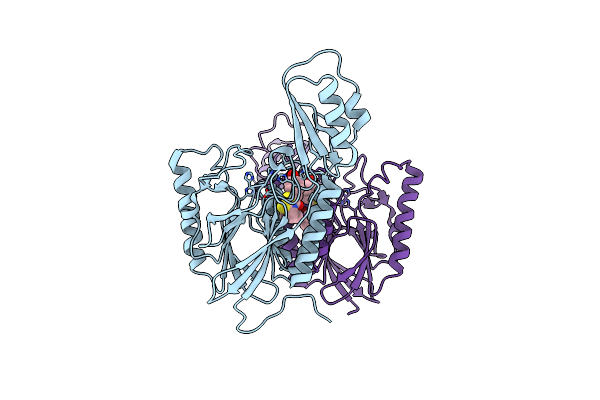 |
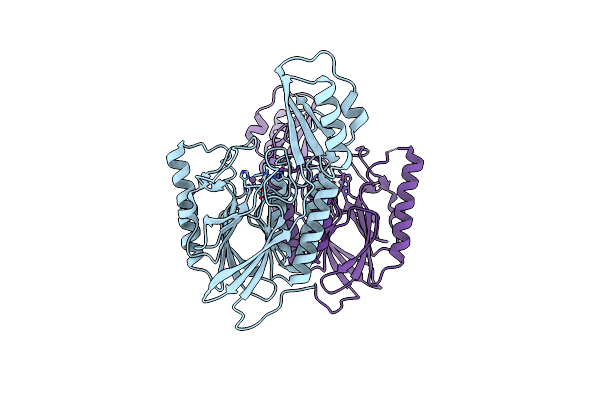
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-01-15 Classification: DNA BINDING PROTEIN/inhibitor Ligands: 2Q0, MN |
 |
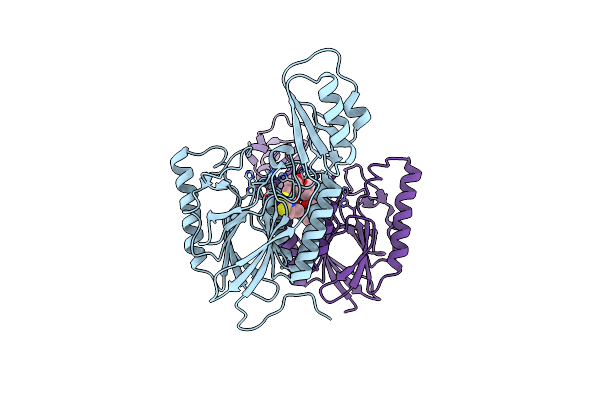
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-01-08 Classification: HYDROLASE Ligands: MN |
 |
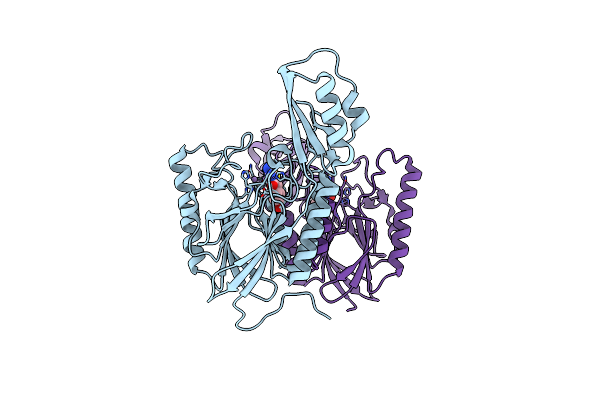
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PW |
 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PK |
 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PV |

