Search Count: 52
 |
Symmetry-Expanded Reconstruction Of Augmin T-Ii Bonsai On The Gtpgammas Microtubule
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CELL CYCLE Ligands: GTP, MG, GSP |
 |
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: CELL CYCLE Ligands: GTP, MG, G2P |
 |
Structure Of A Saccharomyces Cerevisiae Mps1 Peptide Bound To Dwarf Ndc80 Complex
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2024-05-15 Classification: CELL CYCLE Ligands: NI |
 |
Structure Of A Saccharomyces Cerevisiae Ipl1 Peptide Bound To Dwarf Ndc80 Complex
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.95 Å Release Date: 2024-05-15 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: CELL CYCLE Ligands: GTP, MG, GDP, TA1 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: CELL CYCLE |
 |
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: CELL CYCLE |
 |
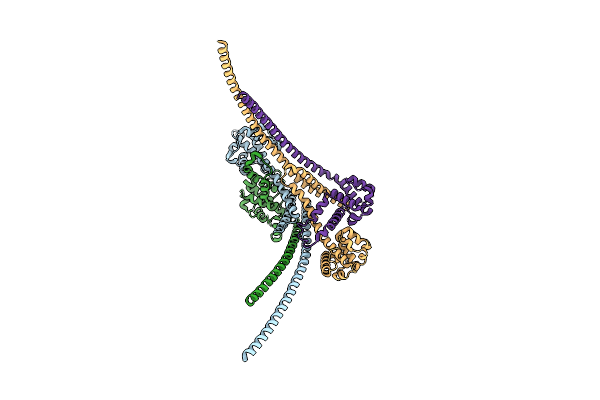
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-03-29 Classification: CELL CYCLE |
 |
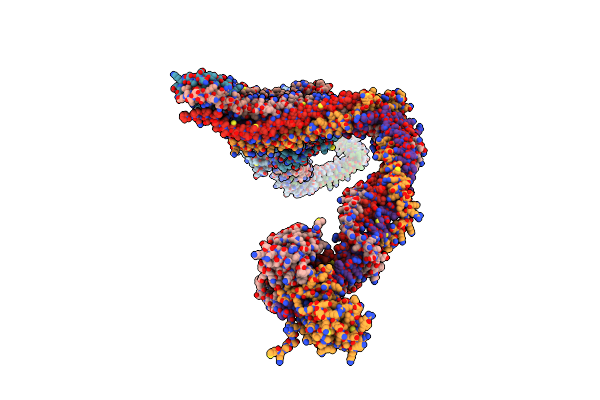
Crystal Structure Of The Yeast Ndc80:Nuf2 Head Region With A Bound Dam1 Segment
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2023-03-29 Classification: CELL CYCLE |
 |
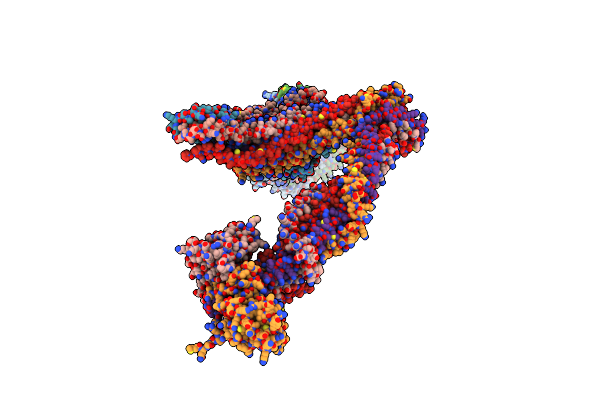
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: CELL CYCLE |
 |
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: CELL CYCLE |
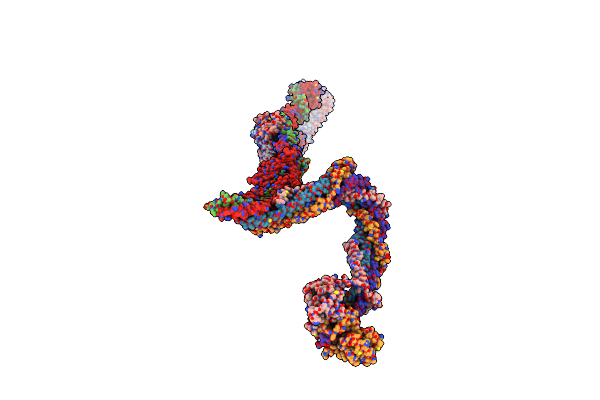 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-09-21 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2020-11-11 Classification: CELL CYCLE Ligands: SO4 |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2016-11-23 Classification: REPLICATION Ligands: MG |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:7.53 Å Release Date: 2016-11-16 Classification: REPLICATION Ligands: HG |
 |
Structure Of The Mind Complex Shows A Regulatory Focus Of Yeast Kinetochore Assembly
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-11-09 Classification: CELL CYCLE |
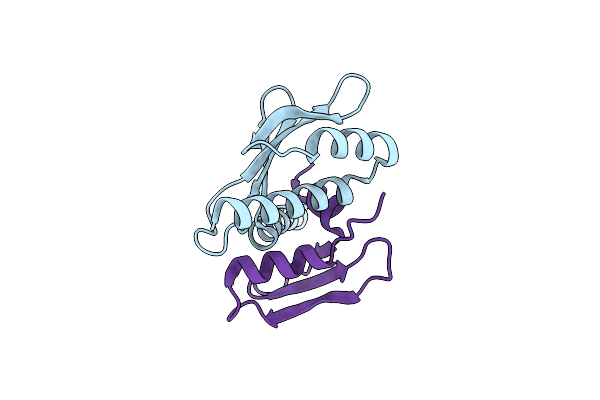 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2013-04-03 Classification: CELL CYCLE |
 |
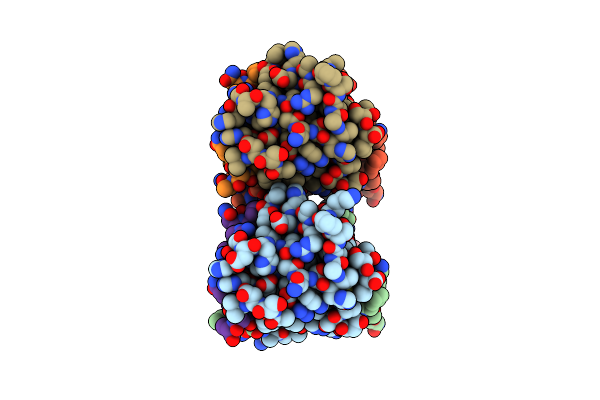
Crystal Structure Of The Chicken Spc24-Spc25 Globular Domain In Complex With Cenp-T Peptide
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-04-03 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2013-01-30 Classification: CELL CYCLE Ligands: GOL |

