Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
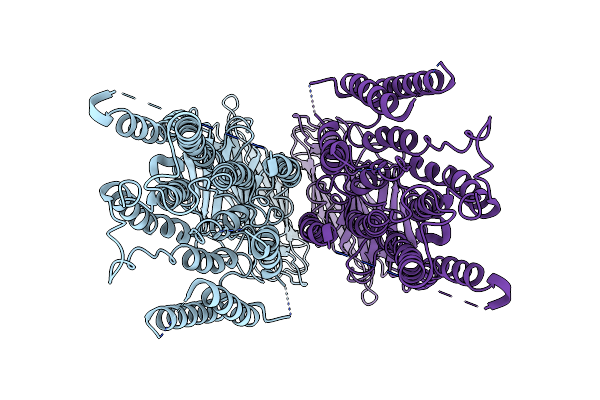
Structure Of The Human Dopamine Transporter In Complex With Beta-Cft, Mrs7292 And Divalent Zinc
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: MEMBRANE PROTEIN Ligands: 42L, A1AAF, NAG, Y01, D10, LNK, HP6, ZN, NA, D12, MYS, OCT |
 |
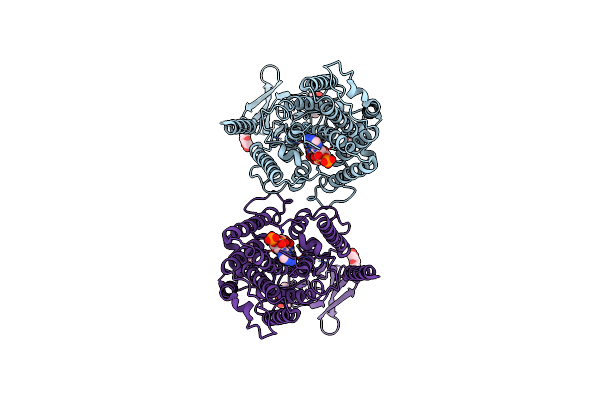
Structure Of The Human Systemic Rnai Defective Transmembrane Protein 1 (Hsidt1)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Aequorea victoria, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSFERASE Ligands: ACO, NAG |
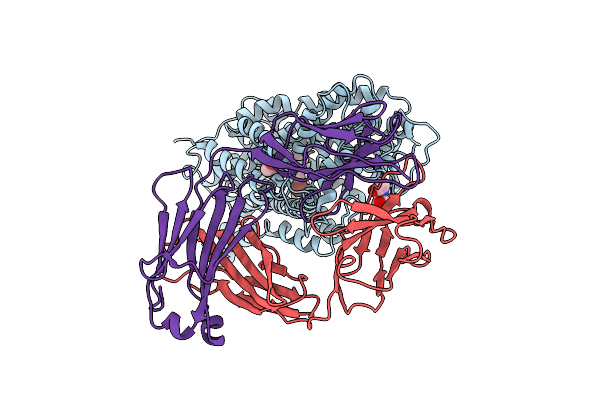 |
Anomalous Bromine Signal Reveals The Position Of Br-Paroxetine Complexed With The Serotonin Transporter At The Central Site
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.70 Å Release Date: 2020-03-25 Classification: TRANSPORT PROTEIN Ligands: NAG, RFS |
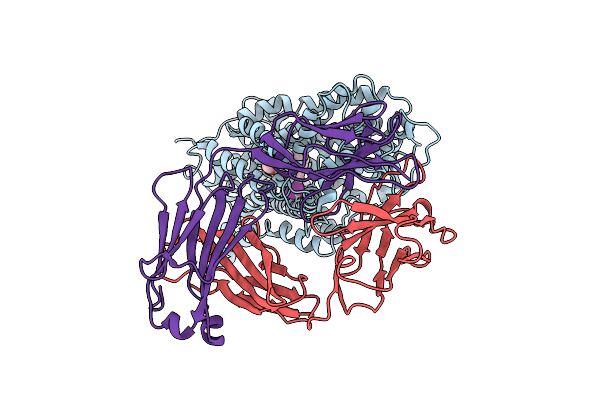 |
Anomalous Iodine Signal Reveals The Position Of I-Paroxetine Complexed With The Serotonin Transporter At The Central Site
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:6.30 Å Release Date: 2020-03-25 Classification: TRANSPORT PROTEIN Ligands: RFY |
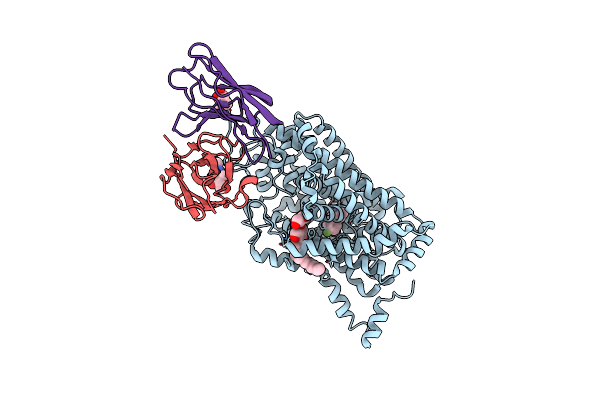 |
Cryo-Em Structure Of The Wild-Type Human Serotonin Transporter Complexed With Paroxetine And 8B6 Fab
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-03-11 Classification: TRANSPORT PROTEIN Ligands: NAG, 8PR, CL, LMT |
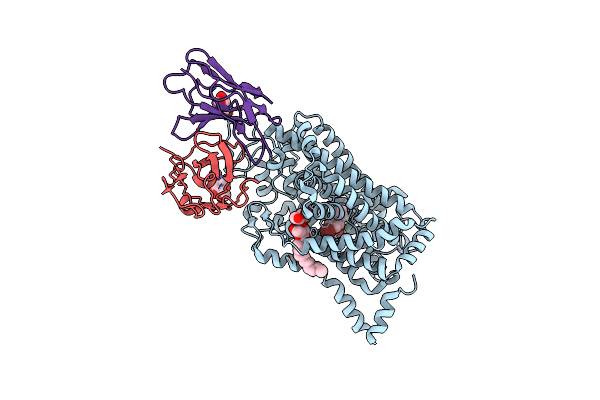 |
Cryo-Em Structure Of The Wild-Type Human Serotonin Transporter Complexed With Br-Paroxetine And 8B6 Fab
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-03-11 Classification: TRANSPORT PROTEIN Ligands: NAG, LMT, RFS |
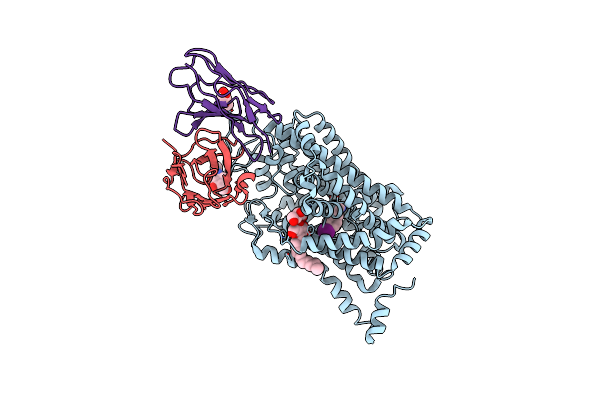 |
Cryo-Em Structure Of The Wild-Type Human Serotonin Transporter Complexed With I-Paroxetine And 8B6 Fab
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-03-11 Classification: TRANSPORT PROTEIN Ligands: NAG, LMT, RFY |
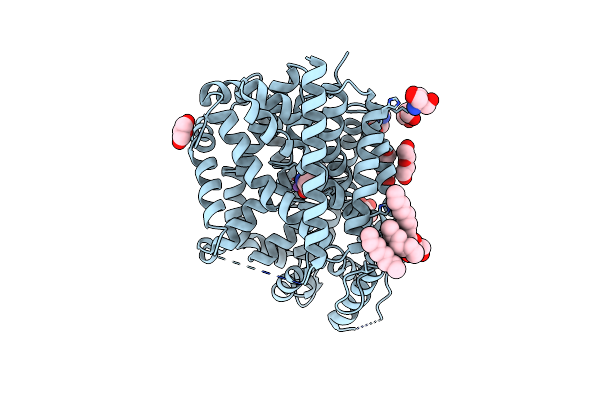 |
Organism: Aquifex aeolicus (strain vf5)
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2020-01-15 Classification: MEMBRANE PROTEIN Ligands: LEU, NA, GOL, PEG, TRS, BOG, OCT |
 |
Crystal Structure Of Dihydrodipicolinate Synthase From Mycobacterium Tuberculosis In Complex With Alpha-Ketopimelic Acid
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2016-08-17 Classification: LYASE |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-05-13 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, PEG, DMS |
 |
Organism: Staphylococcus aureus m1064
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-05-06 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, PEG, DMS |
 |
Organism: Staphylococcus aureus m1064
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-05-06 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, PEG, DMS |
 |
Organism: Staphylococcus aureus m1064
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-05-06 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, LYS, PEG |
 |
Organism: Staphylococcus aureus m1064
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-05-06 Classification: OXIDOREDUCTASE Ligands: ACT, PEG, GOL, DMS |
 |
The Crystal Structure Of Dihydrodipicolinate Reductase From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-08-03 Classification: OXIDOREDUCTASE Ligands: SO4, GOL, ACT |
 |
Crystal Structure Of Penicillin Binding Protein 4 From Staphylococcus Aureus Col In Complex With Cefotaxime
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-11-10 Classification: HYDROLASE/Antibiotics Ligands: CEW |
 |
Crystal Structure Of Penicillin Binding Protein 4 From Staphylococcus Aureus Col In Complex With Ampicillin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-11-10 Classification: HYDROLASE/Antibiotics Ligands: ZZ7 |