Search Count: 14
 |
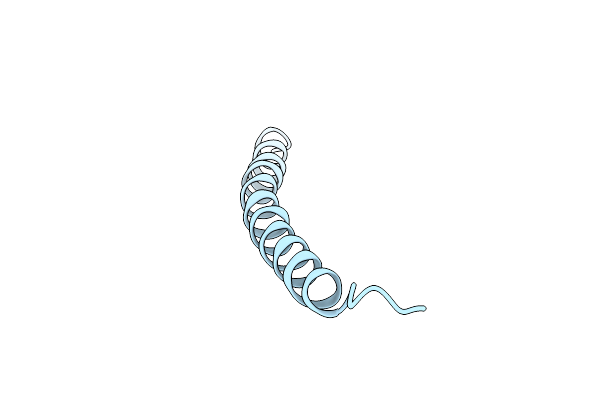
Pf1 Filamentous Bacteriophage: Refinement Of A Molecular Model By Simulated Annealing Using 3.3 Angstroms Resolution X-Ray Fibre Diffraction Data
Organism: Xanthomonas phage xf
Method: FIBER DIFFRACTION Resolution:3.30 Å Release Date: 1996-01-01 Classification: VIRUS |
 |
Pf1 Filamentous Bacteriophage: Refinement Of A Molecular Model By Simulated Annealing Using 3.3 Angstroms Resolution X-Ray Fibre Diffraction Data
Organism: Pseudomonas phage pf1
Method: FIBER DIFFRACTION Resolution:4.00 Å Release Date: 1996-01-01 Classification: VIRUS |
 |
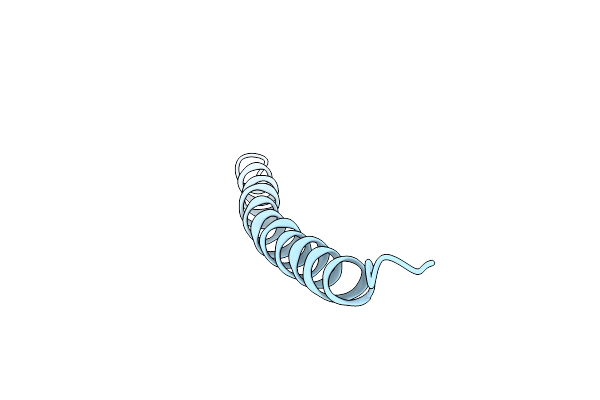
Pf1 Filamentous Bacteriophage: Refinement Of A Molecular Model By Simulated Annealing Using 3.3 Angstroms Resolution X-Ray Fibre Diffraction Data
Organism: Pseudomonas phage pf1
Method: FIBER DIFFRACTION Resolution:3.30 Å Release Date: 1996-01-01 Classification: VIRUS |
 |
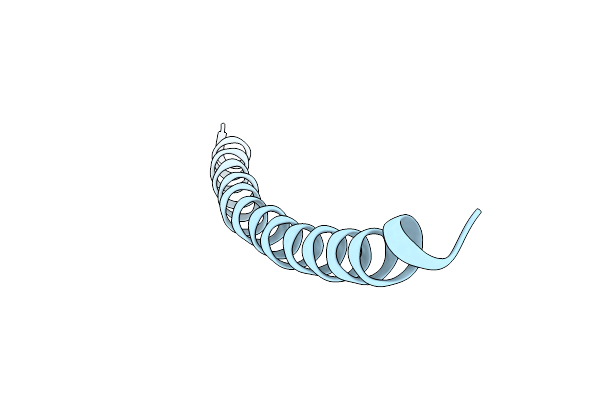
Pf1 Filamentous Bacteriophage: Refinement Of A Molecular Model By Simulated Annealing Using 3.3 Angstroms Resolution X-Ray Fibre Diffraction Data
Organism: Pseudomonas phage pf1
Method: FIBER DIFFRACTION Resolution:3.30 Å Release Date: 1996-01-01 Classification: VIRUS |
 |
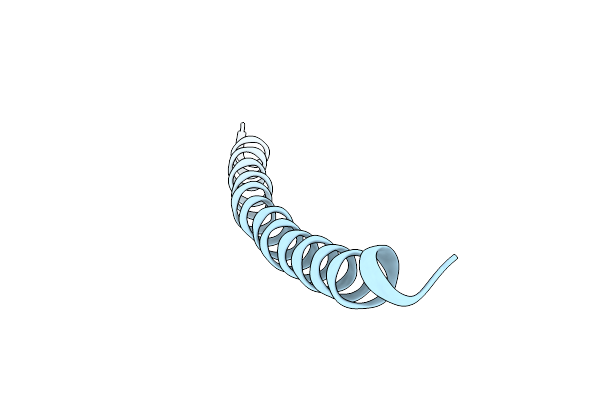
Molecular Models And Structural Comparisons Of Native And Mutant Class I Filamentous Bacteriophages Ff (Fd, F1, M13), If1 And Ike
Organism: Enterobacteria phage fd
Method: FIBER DIFFRACTION Resolution:3.30 Å Release Date: 1994-07-31 Classification: VIRUS |
 |
Molecular Models And Structural Comparisons Of Native And Mutant Class I Filamentous Bacteriophages Ff (Fd, F1, M13), If1 And Ike
Organism: Enterobacteria phage fd
Method: FIBER DIFFRACTION Resolution:3.30 Å Release Date: 1994-07-31 Classification: VIRUS |
 |
Molecular Models And Structural Comparisons Of Native And Mutant Class I Filamentous Bacteriophages Ff (Fd, F1, M13), If1 And Ike
Organism: Enterobacteria phage if1
Method: FIBER DIFFRACTION Resolution:5.00 Å Release Date: 1994-07-31 Classification: VIRUS |
 |
Molecular Models And Structural Comparisons Of Native And Mutant Class I Filamentous Bacteriophages Ff (Fd, F1, M13), If1 And Ike
Organism: Enterobacteria phage ike
Method: FIBER DIFFRACTION Resolution:5.00 Å Release Date: 1994-07-31 Classification: VIRUS |
 |
Two Forms Of Pf1 Inovirus: X-Ray Diffraction Studies On A Structural Phase Transition And A Calculated Libration Normal Mode Of The Asymmetric Unit
Organism: Pseudomonas phage pf1
Method: FIBER DIFFRACTION Resolution:3.30 Å Release Date: 1994-07-31 Classification: VIRUS |
 |
Two Forms Of Pf1 Inovirus: X-Ray Diffraction Studies On A Structural Phase Transition And A Calculated Libration Normal Mode Of The Asymmetric Unit
Organism: Pseudomonas phage pf1
Method: FIBER DIFFRACTION Resolution:4.00 Å Release Date: 1994-07-31 Classification: VIRUS |
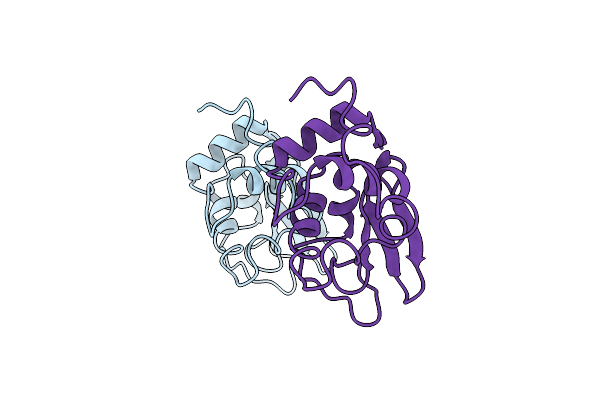 |
Comparison Of Radiation-Induced Decay And Structure Refinement From X-Ray Data Collected From Lysozyme Crystals At Low And Ambient Temperatures
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) |
 |
Comparison Of Radiation-Induced Decay And Structure Refinement From X-Ray Data Collected From Lysozyme Crystals At Low And Ambient Temperatures
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) |
 |
Comparison Of Radiation-Induced Decay And Structure Refinement From X-Ray Data Collected From Lysozyme Crystals At Low And Ambient Temperatures
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) |
 |
Comparison Of Radiation-Induced Decay And Structure Refinement From X-Ray Data Collected From Lysozyme Crystals At Low And Ambient Temperatures
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) |

