Search Count: 14
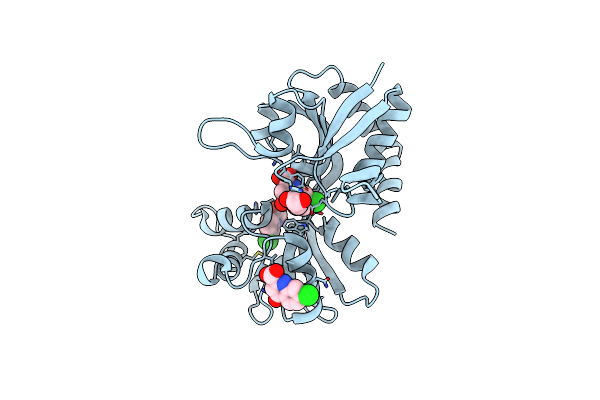 |
Structure Of The Ligand-Binding Domain Of The Ionotropic Glutamate Receptor-Like Glud2 In Complex With 7-Cka
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-12-30 Classification: SIGNALING PROTEIN Ligands: CKA, GOL, CL |
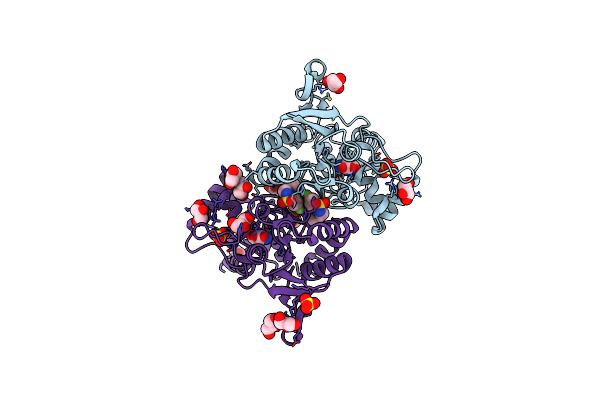 |
Crystal Structure Of The Glua2 Ligand-Binding Domain (S1S2J-L483Y-N754S) In Complex With Glutamate And Bpam-97 At 1.95 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-09-21 Classification: MEMBRANE PROTEIN Ligands: GLU, SO4, CL, GOL, 3TJ |
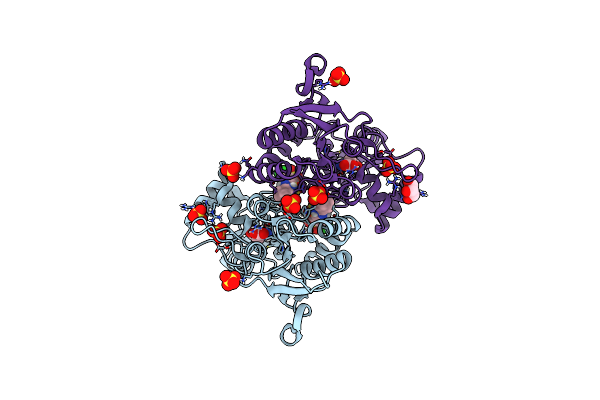 |
Crystal Structure Of The Glua2 Ligand-Binding Domain (S1S2J-L483Y-N754S) In Complex With Glutamate And Cyclothiazide At 1.45 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2011-09-21 Classification: MEMBRANE PROTEIN Ligands: GLU, CYZ, SO4, GOL |
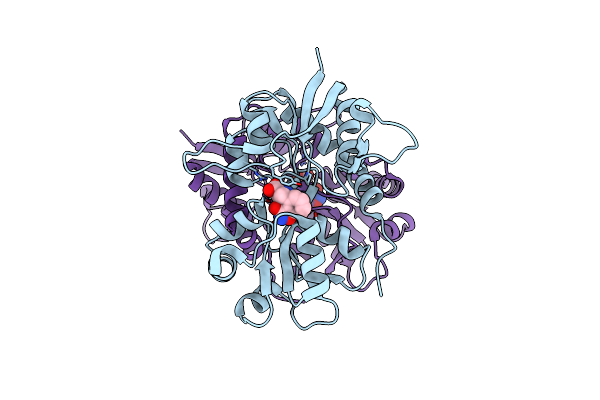 |
Crystal Structure Of The Ligand-Binding Core Of Glur5 In Complex With The Agonist 4-Ahcp
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-07-21 Classification: MEMBRANE PROTEIN Ligands: CL, IBC |
 |
X-Ray Structure Of Iglur5 Ligand-Binding Core (S1S2) In Complex With Dysiherbaine At 1.35A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2009-03-17 Classification: MEMBRANE PROTEIN Ligands: DYH, SO4, GOL, CL |
 |
X-Ray Structure Of Iglur5 Ligand-Binding Core (S1S2) In Complex With Msviii-19 At 2.10A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-03-17 Classification: MEMBRANE PROTEIN Ligands: MS8 |
 |
X-Ray Structure Of Iglur4 Flip Ligand-Binding Core (S1S2) In Complex With (S)-Glutamate At 1.40A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2008-12-09 Classification: MEMBRANE PROTEIN Ligands: GLU, SO4, GOL |
 |
X-Ray Structure Of Iglur4 Flip Ligand-Binding Core (S1S2) In Complex With (S)-Ampa At 1.90A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-12-09 Classification: MEMBRANE PROTEIN Ligands: AMQ, SO4, GOL, ACY |
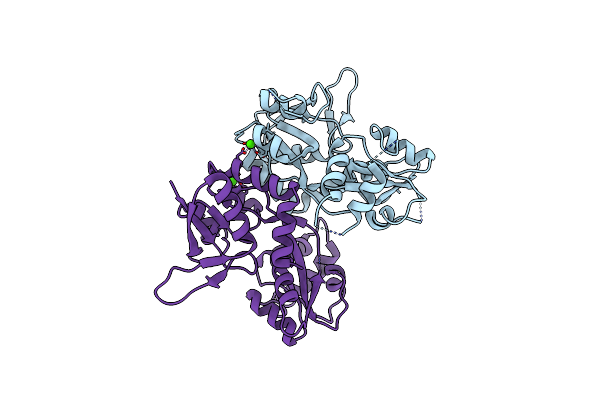 |
Structure Of The Ligand-Binding Core Of The Ionotropic Glutamate Receptor-Like Glurdelta2 In The Apo Form
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2007-08-07 Classification: RECEPTOR Ligands: CA |
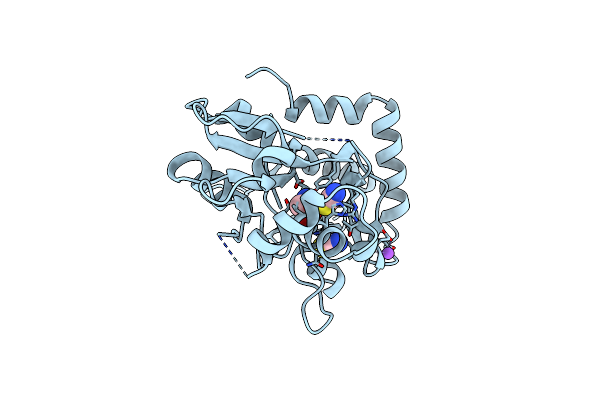 |
Structure Of The Ligand-Binding Core Of The Ionotropic Glutamate Receptor-Like Glurdelta2 In Complex With D-Serine
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2007-08-07 Classification: RECEPTOR Ligands: DSN, NA, CL, SCN |
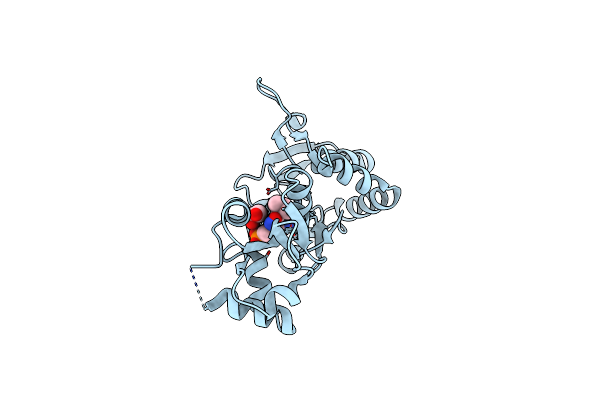 |
Crystal Structure Of The Ligand-Binding Core Of Iglur5 In Complex With The Antagonist (S)-Atpo At 1.85 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-07-03 Classification: MEMBRANE PROTEIN Ligands: AT1, GOL |
 |
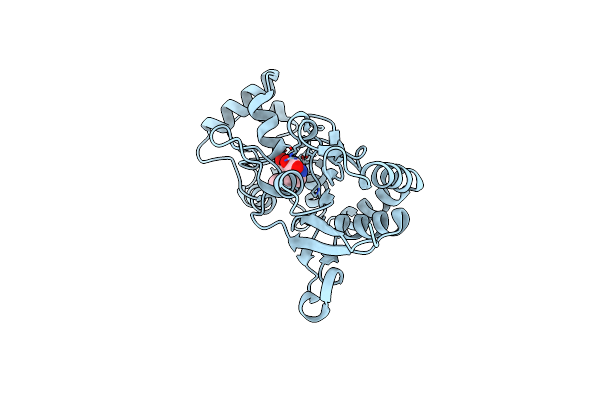
Crystal Structure Of The Ligand-Binding Core Of Iglur5 In Complex With The Partial Agonist Domoic Acid At 2.5 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-07-03 Classification: MEMBRANE PROTEIN Ligands: DOQ |
 |
Crystal Structure Of The Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant In Complex With The Partial Agonist Kainic Acid At 2.1 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-08-30 Classification: MEMBRANE PROTEIN Ligands: KAI |
 |
Crystal Structure Of The Kainate Receptor Glur5 Ligand-Binding Core In Complex With (S)-Glutamate
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2005-02-01 Classification: MEMBRANE PROTEIN Ligands: SO4, GLU |

