Search Count: 18
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: ZN, NDG, NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: ZN, NDG, NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: NAG, YCP, CL |
 |
Structure Of Human Sit1:Ace2 Complex (Open Pd Conformation) Bound To L-Pipecolate
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: ZN, NDG, NAG, YCP, CL |
 |
Structure Of Human Sit1:Ace2 Complex (Closed Pd Conformation) Bound To L-Pipecolate
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: MEMBRANE PROTEIN Ligands: ZN, NDG, NAG, YCP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: MEMBRANE PROTEIN Ligands: NAG, QUS, Y01 |
 |
Thermostabilised Full Length Human Mglur5-5M With Orthosteric Antagonist, Ly341495
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: MEMBRANE PROTEIN Ligands: NAG, Z99 |
 |
Thermostabilised 7Tm Domain Of Human Mglu5 Receptor Bound To Photoswitchable Ligand Alloswitch-1
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2021-09-08 Classification: SIGNALING PROTEIN Ligands: 4YI |
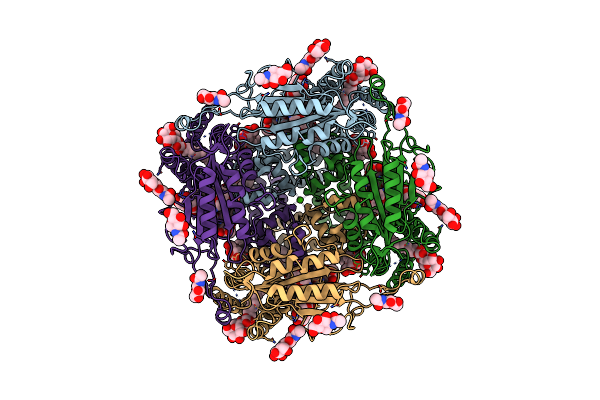 |
Cryoem Structure Of Human Polycystin-2/Pkd2 In Udm Supplemented With Pi(4,5)P2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-11-20 Classification: MEMBRANE PROTEIN Ligands: CLR, NAG, UMQ, CA |
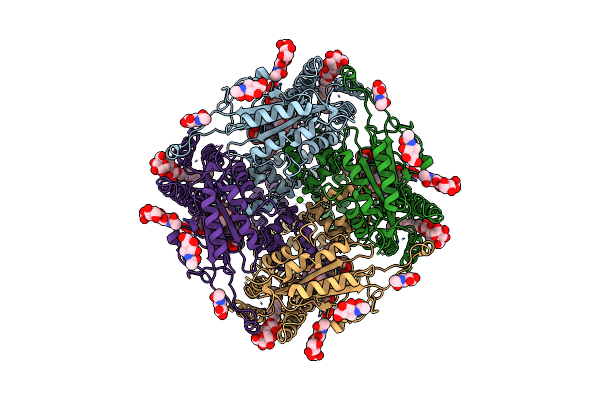 |
Cryoem Structure Of Human Polycystin-2/Pkd2 In Udm Supplemented With Pi(3,5)P2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-11-20 Classification: MEMBRANE PROTEIN Ligands: NAG, CLR, UMQ, CA |
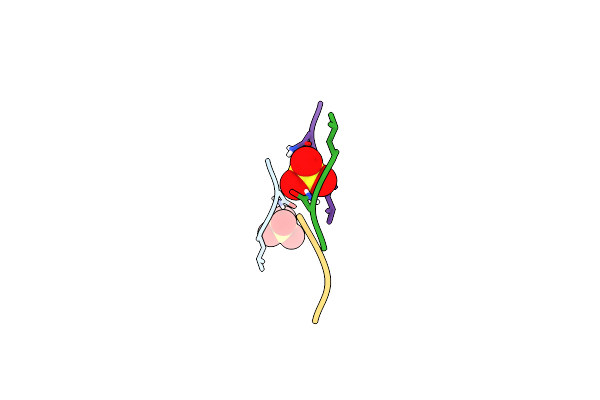 |
Structure Of 283-Lgny-286, The Steric Zipper That Supports The Self-Association Of P. Stuartii Omp-Pst2 Into Dimers Of Trimers
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2018-02-21 Classification: CELL ADHESION Ligands: SO4 |
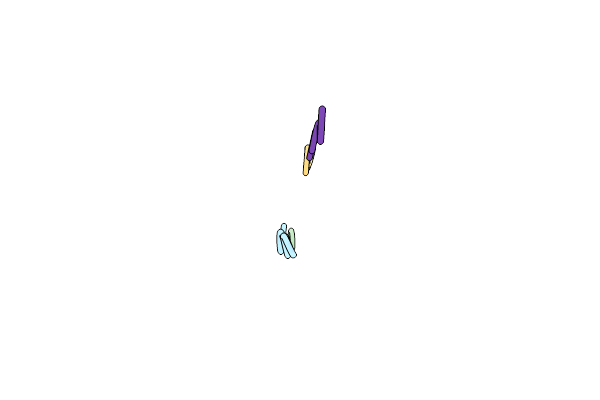 |
Structure Of 206-Gvvtse-211, The Steric Zipper That Supports The Self-Association Of P. Stuartii Omp-Pst1 Into Dimers Of Trimers
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2018-02-21 Classification: CELL ADHESION |
 |
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2018-02-21 Classification: TRANSPORT PROTEIN Ligands: CA |
 |
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-02-21 Classification: CELL ADHESION Ligands: LDA, CA, CL |
 |
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-02-21 Classification: CELL ADHESION Ligands: LDA, CA |
 |
Structure Of Porin Omp-Pst1 From P. Stuartii; The Crystallographic Symmetry Generates A Dimer Of Trimers.
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-03-09 Classification: TRANSPORT PROTEIN Ligands: CA |
 |
Structure Of Porin Omp-Pst2 From P. Stuartii; The Asymmetric Unit Contains A Dimer Of Trimers.
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-09 Classification: TRANSPORT PROTEIN Ligands: LDA, FTT, MYR, SO4 |

