Search Count: 63
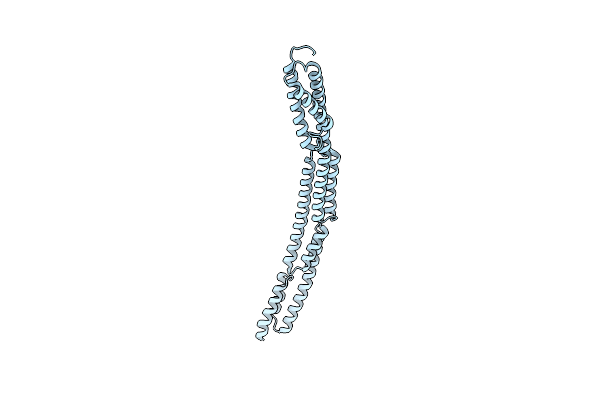 |
Novel Structure Of The N-Terminal Helical Domain Of Biba, A Group B Streptococcus Immunogenic Bacterial Adhesin
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2020-08-12 Classification: IMMUNE SYSTEM |
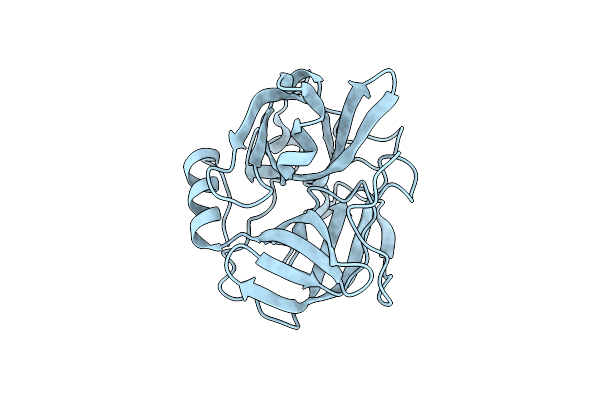 |
Structure Of N-Terminus Locked Esp With Eight Pro-Peptide Residues - V67C, D255C
Organism: Staphylococcus epidermidis (strain atcc 12228)
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2019-09-04 Classification: HYDROLASE |
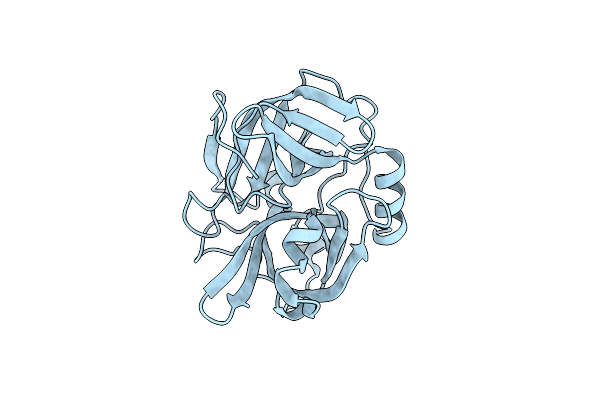 |
Structure Of N-Terminus Locked Esp With One Pro-Peptide Residue - V67C, D255C
Organism: Staphylococcus epidermidis (strain atcc 12228)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2019-08-28 Classification: HYDROLASE |
 |
Organism: Staphylococcus epidermidis (strain atcc 12228)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-08-21 Classification: HYDROLASE |
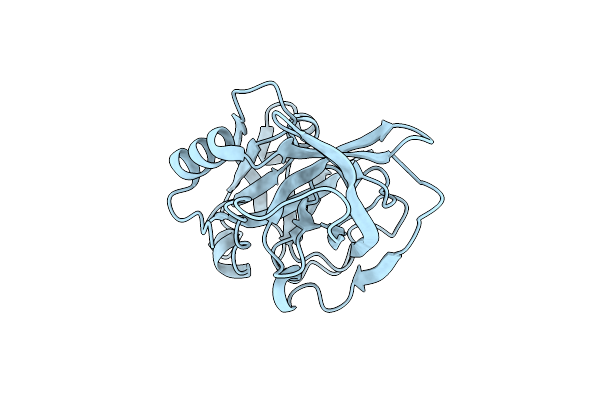 |
Structure Of Active-Site Serine Mutant Of Esp, Serine Protease From Staphylococcus Epidermidis
Organism: Staphylococcus epidermidis (strain atcc 12228)
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2019-08-14 Classification: HYDROLASE |
 |
Organism: Staphylococcus epidermidis (strain atcc 12228)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-08-14 Classification: HYDROLASE |
 |
Crystal Structure Of The N-Terminal Domain Of Fibronectin- Binding Protein Pava From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-07-31 Classification: CELL ADHESION |
 |
Organism: Staphylococcus epidermidis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-09-04 Classification: HYDROLASE |
 |
Structural Analysis Of Adhesive Tip Pilin, Gbs104 From Group B Streptococcus Agalactiae
Organism: Streptococcus agalactiae serogroup v
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-03-27 Classification: CELL ADHESION Ligands: EPE, MG |
 |
Structural Analysis Of Adhesive Tip Pilin, Gbs104 From Group B Streptococcus Agalactiae
Organism: Streptococcus agalactiae serogroup v
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-03-27 Classification: CELL ADHESION Ligands: MG, ACT |
 |
Structural Analysis Of Adhesive Tip Pilin, Gbs104 From Group B Streptococcus Agalactiae
Organism: Streptococcus agalactiae serogroup v
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2013-03-27 Classification: CELL ADHESION Ligands: MG, CD, LI |
 |
Organism: Streptococcus agalactiae serogroup v
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2011-10-26 Classification: HYDROLASE |
 |
The Crystal Structure Of The Complex Of Streptococcus Agalactiae Sortase C1 And Mtset
Organism: Streptococcus agalactiae serogroup v
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2011-10-26 Classification: HYDROLASE Ligands: ETM, SO4, CL |
 |
Organism: Streptococcus agalactiae serogroup v
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-09-07 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Streptococcus agalactiae serogroup v
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-09-07 Classification: HYDROLASE |
 |
Organism: Streptococcus agalactiae serogroup v
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2011-09-07 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Streptococcus agalactiae serogroup v
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2011-09-07 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Actinomyces naeslundii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-08-24 Classification: CELL ADHESION Ligands: ZN |
 |
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-05-04 Classification: CELL ADHESION/BLOOD CLOTTING |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2011-05-04 Classification: CELL ADHESION Ligands: MG |

