Search Count: 23
 |
Structural And Biochemical Rationale For Beta Variant Protein Booster Vaccine Broad Cross-Neutralization Of Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structural And Biochemical Rationale For Beta Variant Protein Booster Vaccine Broad Cross-Neutralization Of Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Trka Jm-Kinase With 1-(9{H}-Fluoren-9-Yl)-3-(2-Methyl-4-Phenyl-Pyrimidin-5-Yl)Urea
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2016-12-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 6UE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2016-12-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 6UF |
 |
Trka Jm-Kinase With 2-Fluoro-{N}-[2-(4-Fluorophenyl)-6-Methyl-3-Pyridyl]-4-(Trifluoromethyl)Benzamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2016-12-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 6UG |
 |
Trka Jm-Kinase With 1-(5-Methyl-3-Phenyl-1,2-Oxazol-4-Yl)-3-[[2-(Trifluoromethyl)Phenyl]Methyl]Urea
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2016-12-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 6UH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2016-12-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 6UJ |
 |
Trka Jm-Kinase With 1-(2-Methyl-4-Phenyl-Pyrimidin-5-Yl)-3-[[2-(Trifluoromethyl)Phenyl]Methyl]Urea
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2016-12-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 6UK |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2016-12-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 6UM |
 |
Structure-Activity Relationship Guides Enantiomeric Preference Among Potent Inhibitors Of B. Anthracis Dihydrofolate Reductase
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-02-13 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: CA, CL, 34S, 34R |
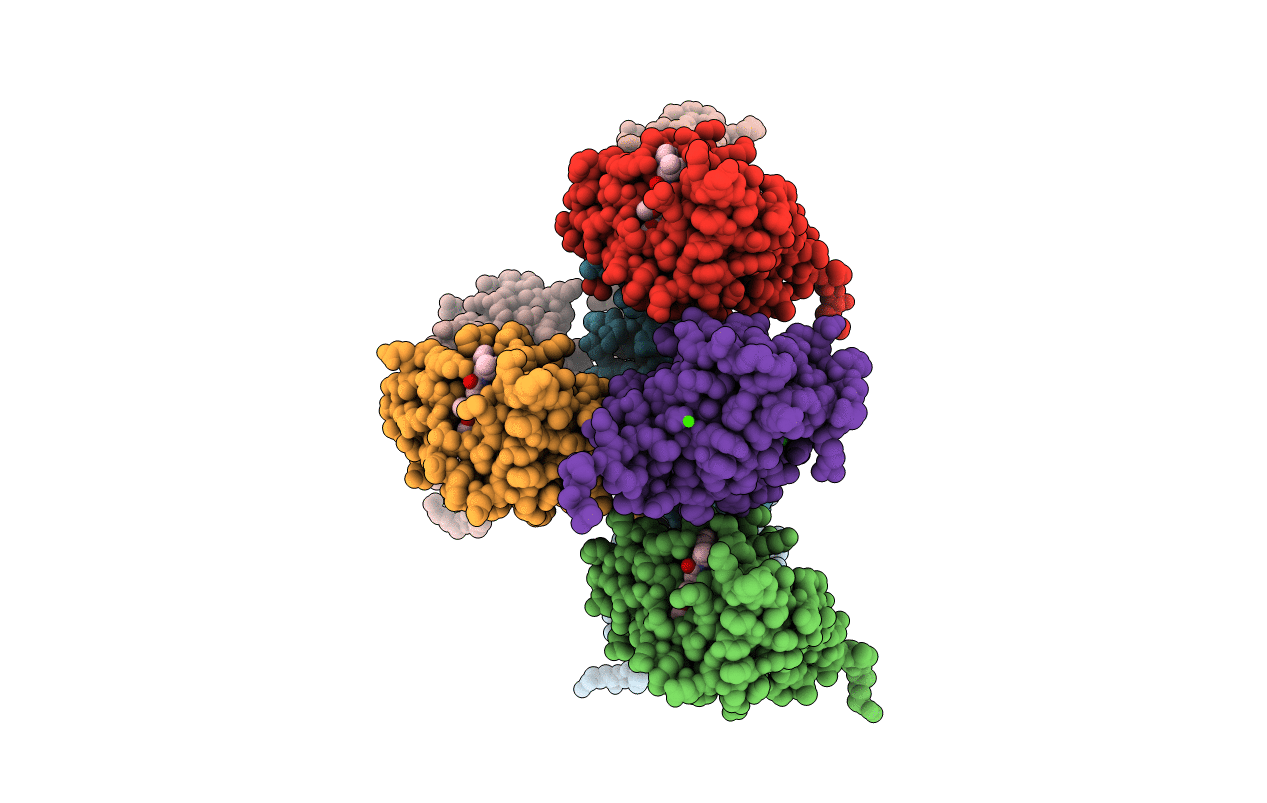 |
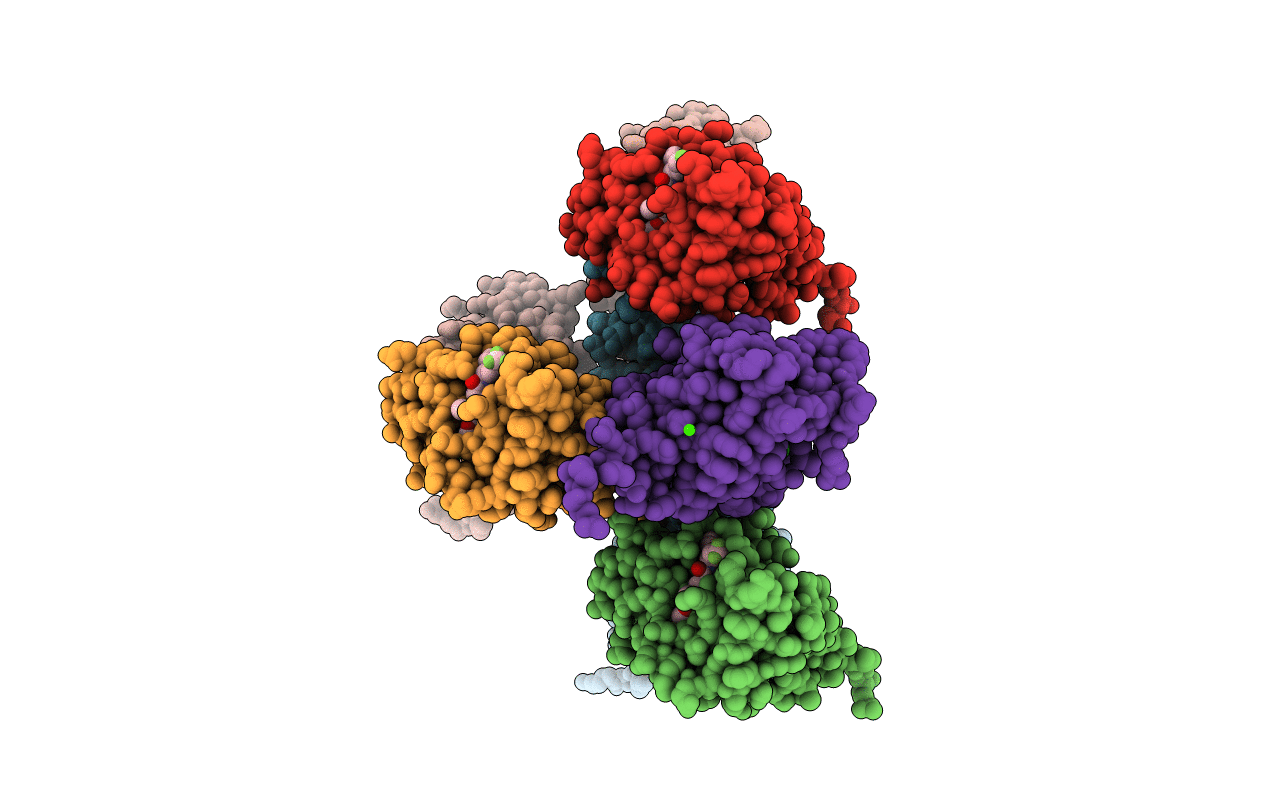
Structure-Activity Relationship Guides Enantiomeric Preference Among Potent Inhibitors Of B. Anthracis Dihydrofolate Reductase
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-02-13 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: CA, CL, 31I |
 |
Structure-Activity Relationship Guides Enantiomeric Preference Among Potent Inhibitors Of B. Anthracis Dihydrofolate Reductase
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-02-13 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: CA, CL, 35I |
 |
Structure-Activity Relationship Guides Enantiomeric Preference Among Potent Inhibitors Of B. Anthracis Dihydrofolate Reductase
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-02-13 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: 52J, CA, CL, 52I |
 |
Structure-Activity Relationship Guides Enantiomeric Preference Among Potent Inhibitors Of B. Anthracis Dihydrofolate Reductase
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-02-13 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: CA, 53I, CL, 53J |
 |
S. Aureus Dihydrofolate Reductase Co-Crystallized With Propyl-Dap Isobutenyl-Dihydrophthalazine Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-01-09 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: 0U5, NAP, GOL |
 |
S. Aureus Dihydrofolate Reductase Co-Crystallized With Ethyl-Dap Isobutenyl-Dihydrophthalazine Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-01-09 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, 0U6, GOL |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-05-20 Classification: HORMONE/SIGNALING PROTEIN Ligands: BR |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-05-20 Classification: HORMONE/SIGNALING PROTEIN |
 |
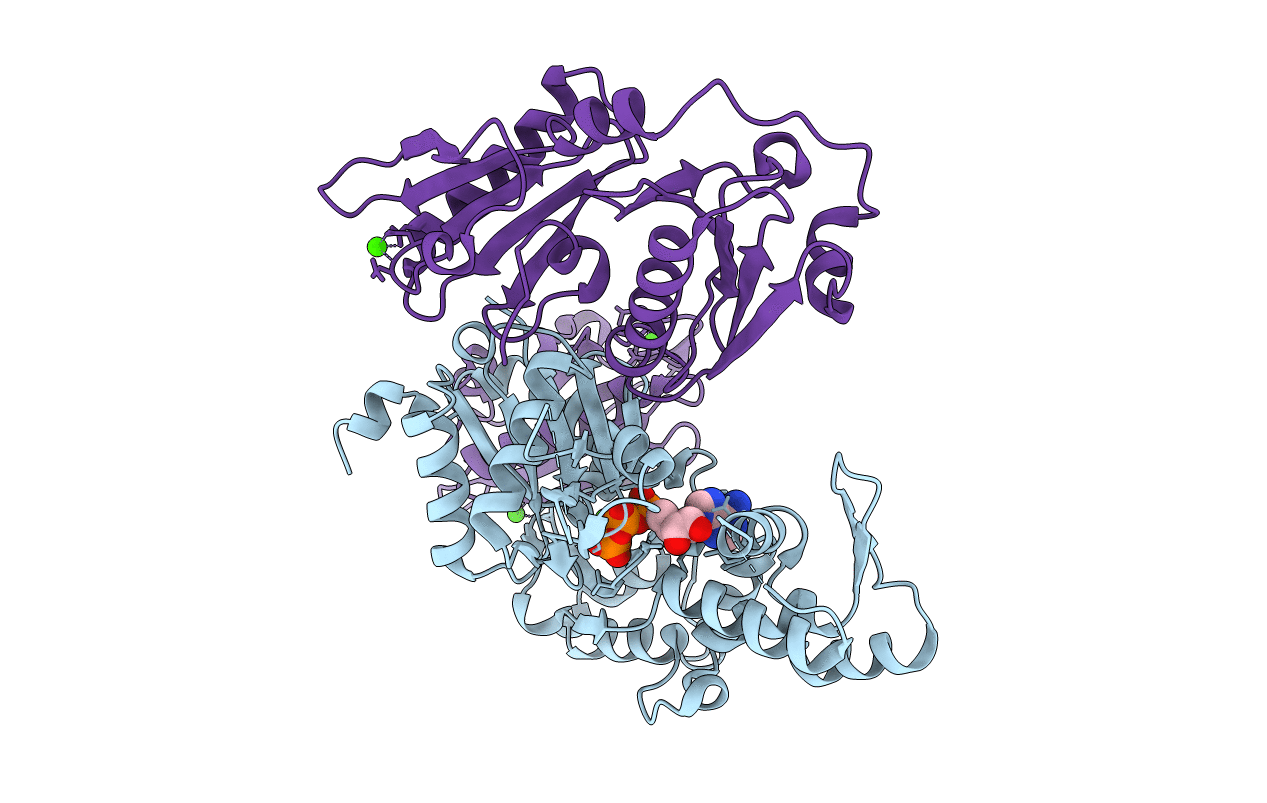
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2008-05-20 Classification: HORMONE/SIGNALING PROTEIN |
 |
Organism: Equus caballus, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2004-07-27 Classification: CONTRACTILE PROTEIN Ligands: CA, ATP |

