Search Count: 17
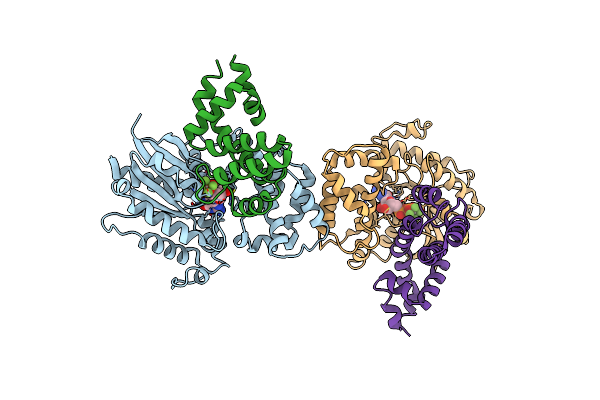 |
Structure Of Human Regulator Of G Protein Signaling 2 (Rgs2) In Complex With Murine Galpha-Q(R183C)
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:7.40 Å Release Date: 2013-01-30 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: GDP, ALF, MG |
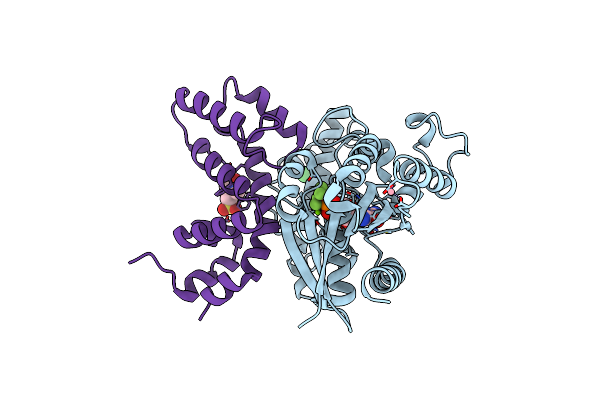 |
Structure Of Human Regulator Of G Protein Signaling 2 (Rgs2) In Complex With Murine Galpha-Q(R183C)
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2013-01-30 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: GDP, ALF, CO, CL, MG, MES |
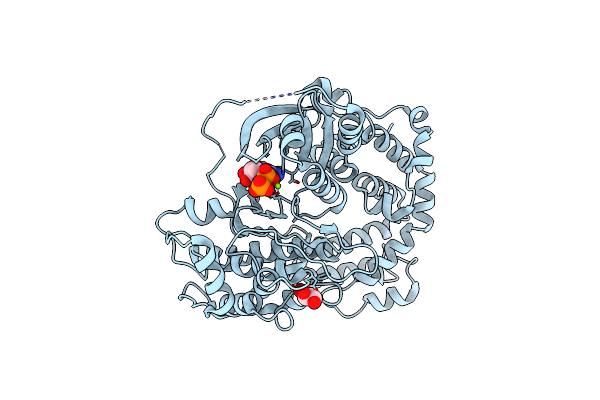 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-06-06 Classification: TRANSFERASE Ligands: ATP, MG, CL, GOL |
 |
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-09-21 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2011-09-21 Classification: HYDROLASE Ligands: CL, NA |
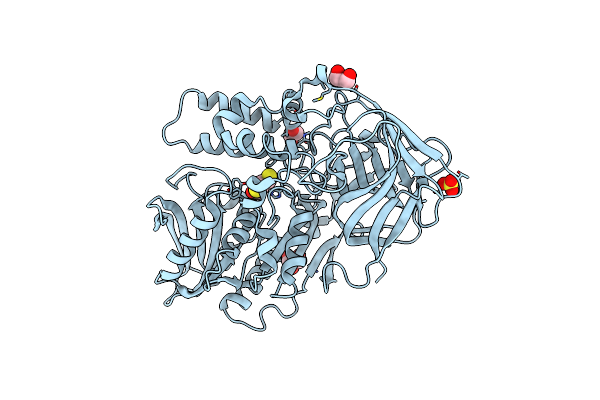 |
Organism: Rhodococcus sp. mb1 'bresler 1999'
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-06-16 Classification: HYDROLASE Ligands: CL, DBC, GOL, SO4 |
 |
Organism: Rhodococcus sp. mb1 'bresler 1999'
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-06-16 Classification: HYDROLASE Ligands: CL, DBC, GOL, SO4 |
 |
Organism: Rhodococcus sp. mb1 'bresler 1999'
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2010-06-16 Classification: HYDROLASE Ligands: CL, DBC, GOL, SO4 |
 |
Organism: Rhodococcus sp. mb1 'bresler 1999'
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2010-06-16 Classification: HYDROLASE Ligands: CL, DBC, GOL, SO4 |
 |
Organism: Rhodococcus sp. mb1 'bresler 1999'
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2010-06-16 Classification: HYDROLASE Ligands: CL, GOL, SO4 |
 |
Organism: Rhodococcus sp. mb1 'bresler 1999'
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2010-06-16 Classification: HYDROLASE Ligands: CL, DBC, GOL, SO4 |
 |
Thermostable Cocaine Esterase With Mutations L169K And G173Q, Bound To Dtt Adduct
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-03-02 Classification: HYDROLASE Ligands: CL, DBC, GOL |
 |
Organism: Rattus norvegicus, Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2008-01-15 Classification: SIGNALING PROTEIN complex Ligands: ALF, MG, GDP |
 |
Organism: Spodoptera frugiperda
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-12-19 Classification: cell adhesion, Membrane Protein Ligands: CL, URE, GOL |
 |
Moesin From Spodoptera Frugiperda Reveals The Coiled-Coil Domain At 3.0 Angstrom Resolution
Organism: Spodoptera frugiperda
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2006-12-19 Classification: Cell Adhesion, Membrane Protein Ligands: CL, URE, GOL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2005-11-15 Classification: SIGNALING PROTEIN Ligands: MG, ALF, GDP |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-11-15 Classification: SIGNALING PROTEIN Ligands: GDP |

