Search Count: 10
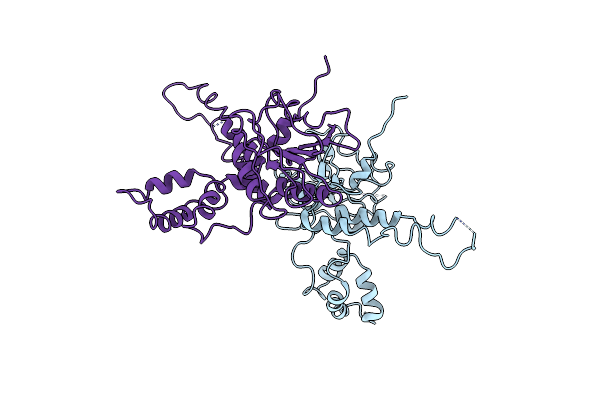 |
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2023-12-06 Classification: DNA BINDING PROTEIN |
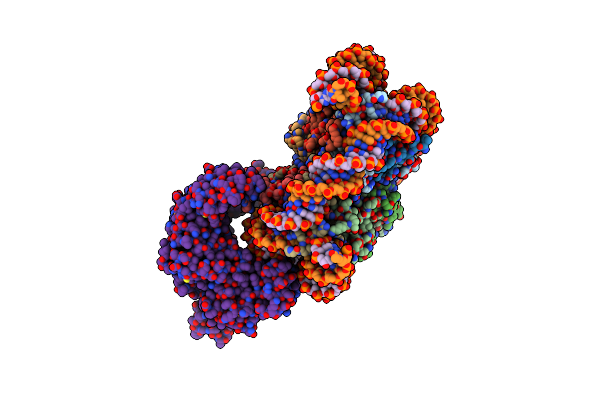 |
Cryo-Em Structure Of Fft3-Nucleosome Complex With Fft3 Bound To Shl+2 Position Of The Nucleosome
Organism: Drosophila melanogaster, Schizosaccharomyces pombe 972h-, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: DNA BINDING PROTEIN |
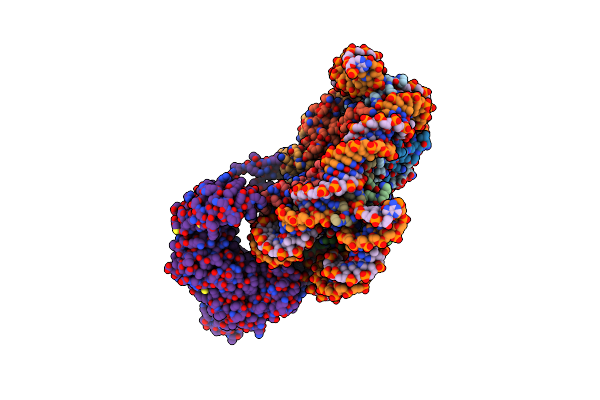 |
Cryo-Em Structure Of Fft3-Nucleosome Complex With Fft3 Bound To Shl+3 Position Of The Nucleosome
Organism: Drosophila melanogaster, Schizosaccharomyces pombe 972h-, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: DNA BINDING PROTEIN |
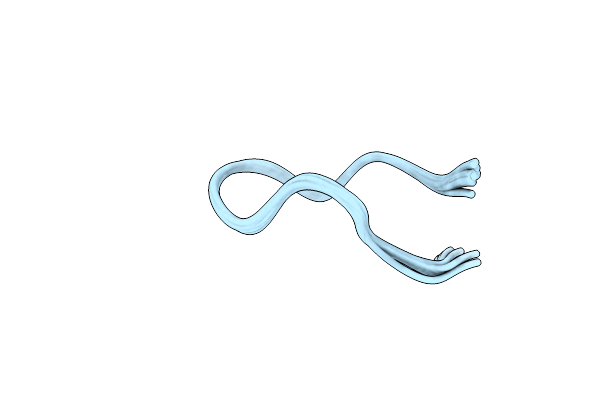 |
Structure Of The Globular Isoform Of The Novel Conotoxin Pnid Derived From Conus Pennaceus
|
 |
Structure Of The Ribbon Isoform Of The Novel Conotoxin Pnid Derived From Conus Pennaceus
|
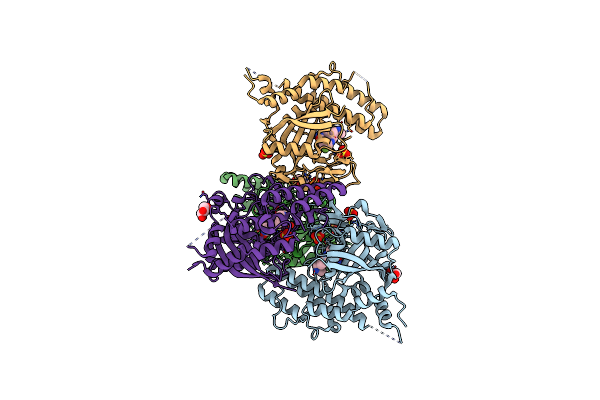 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-06-17 Classification: TRANSFERASE/INHIBITOR Ligands: RPB, GOL, SO4 |
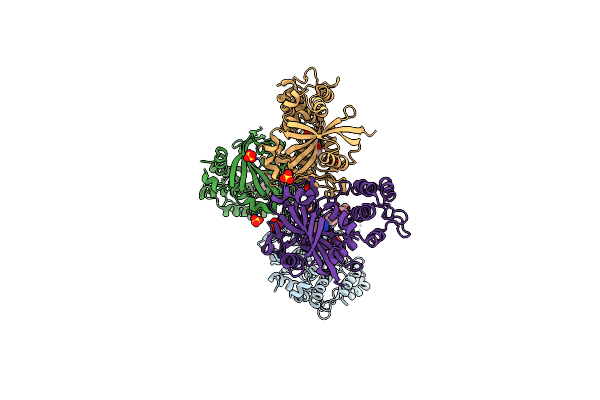 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-06-17 Classification: TRANSFERASE/INHIBITOR Ligands: L1S, SO4 |
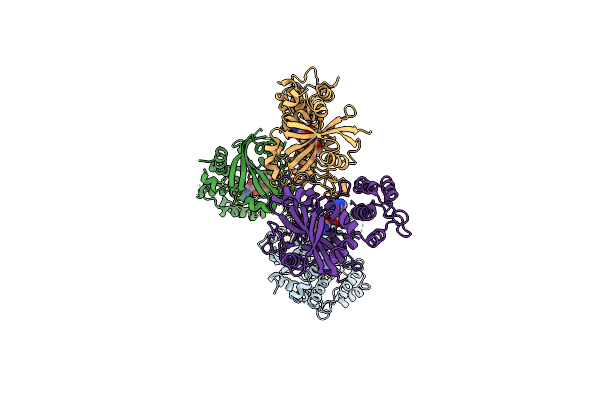 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-06-17 Classification: TRANSFERASE/INHIBITOR Ligands: UHB, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-05 Classification: transferase/transferase inhibitor Ligands: DMS, 1PE, L1S, SO4 |
 |
Organism: Legionella pneumophila subsp. pneumophila (strain philadelphia 1 / atcc 33152 / dsm 7513)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-11-06 Classification: HYDROLASE Ligands: SO4, GOL, ZN, ACT |

