Search Count: 80
 |
Cryo Em Structure Of Kca3.1_R355K_I/Calmodulin Channel In Complex With Rimtuzalcap
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN Ligands: K, A1B92, CA |
 |
Cryo Em Structure Of Kca3.1_R355K_Ii/Calmodulin Channel In Complex With Rimtuzalcap
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: TRANSPORT PROTEIN Ligands: K |
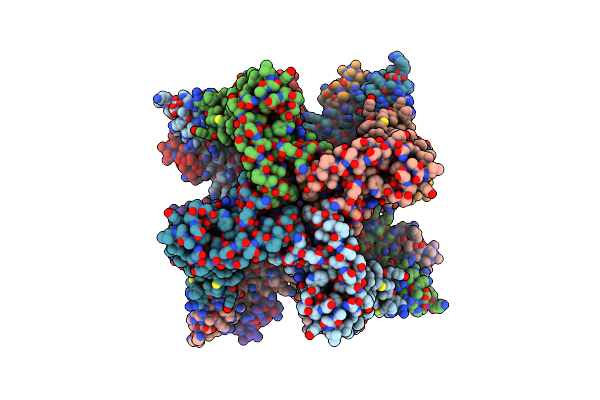 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: K, 1KP, CA |
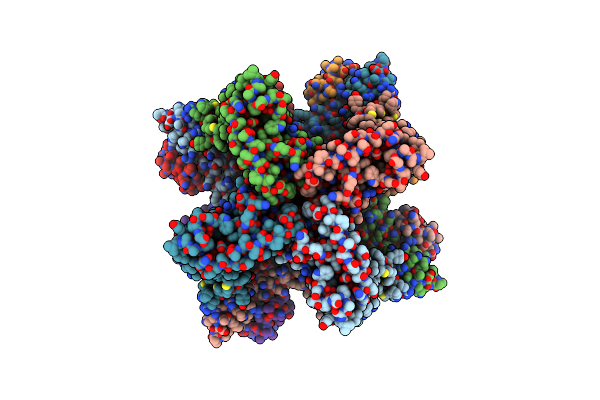 |
Cryo-Em Structure Of Kca2.2_I/Calmodulin Channel In Complex With Rimtuzalcap
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: K, A1B92, CA |
 |
Cryo-Em Structure Of Kca2.2_Ii/Calmodulin Channel In Complex With Rimtuzalcap
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: K, A1B92 |
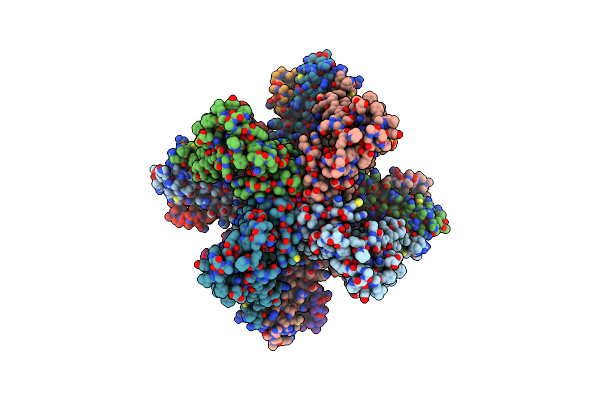 |
Organism: Homo sapiens, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: K, 1KP, CA |
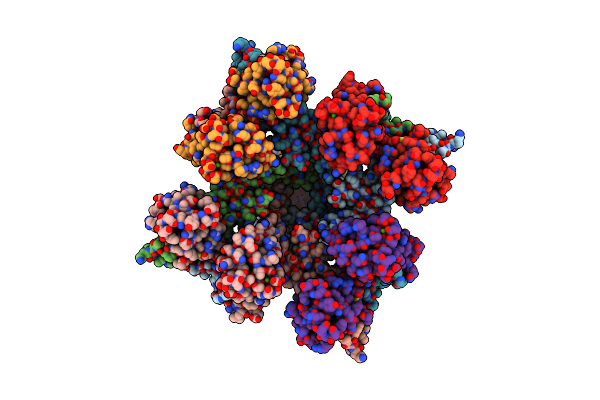 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: K, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: K, Y7Z, CA |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Resolution:3.62 Å Release Date: 2025-04-23 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Cryo-Em Structure Of The Human Kca3.1/Calmodulin Channel In Complex With Ca2+ And 1,4-Dihydropyridine (Dhp-103)
Organism: Homo sapiens, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Crystal Structure Of Adenosylcobinamide Kinase / Adenosylcobinamide Phosphate Guanylyltransferase From Methylocapsa Palsarum
Organism: Methylocapsa palsarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Adenosylcobinamide Kinase / Adenosylcobinamide Phosphate Guanylyltransferase Complexed With Adenosylcobinamide-Phosphate
Organism: Methylocapsa palsarum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: B12, 5AD, PO4 |
 |
Adenosylcobinamide Kinase/Adenosylcobinamide Phosphate Guanylyltransferase -C91S
Organism: Methylocapsa palsarum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Adenosylcobinamide Kinase / Adenosylcobinamide Phosphate Guanylyltransferase Complexed With Gdp
Organism: Methylocapsa palsarum
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: GDP, DPO |
 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
 |
Crystal Structure Of Sulfur Transferase From Frondihabitans Sp. Pamc28461 Crystallized In The P1 Space Group
Organism: Frondihabitans sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-24 Classification: TRANSFERASE |

