Search Count: 216
 |
Organism: Montipora sp. 20
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-23 Classification: FLUORESCENT PROTEIN |
 |
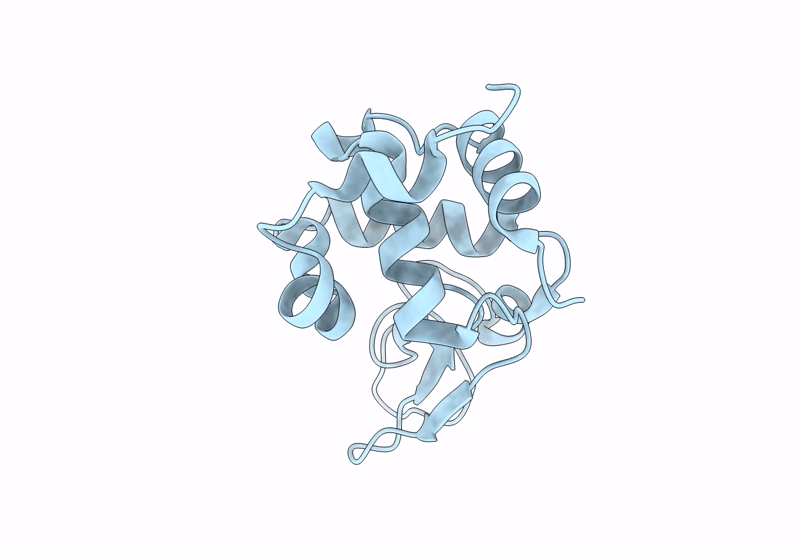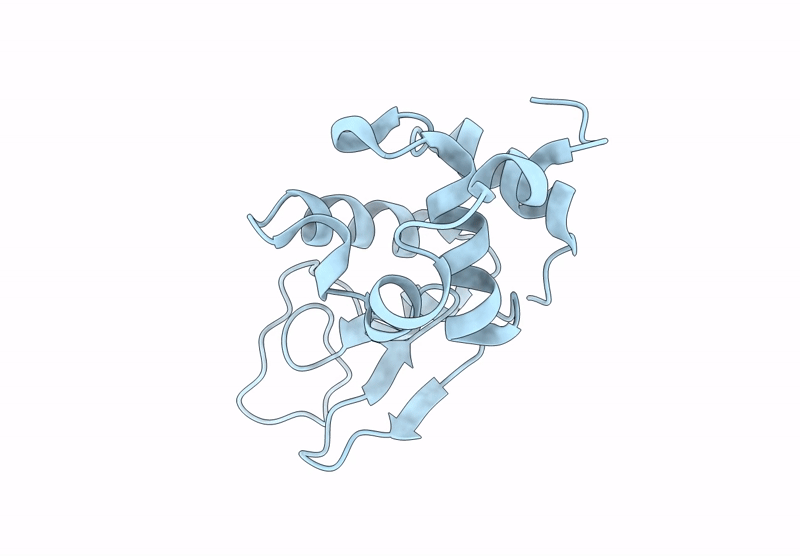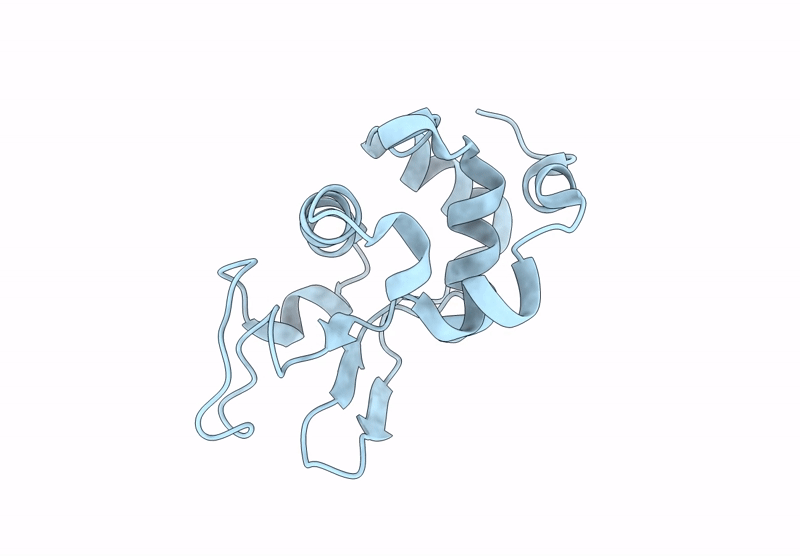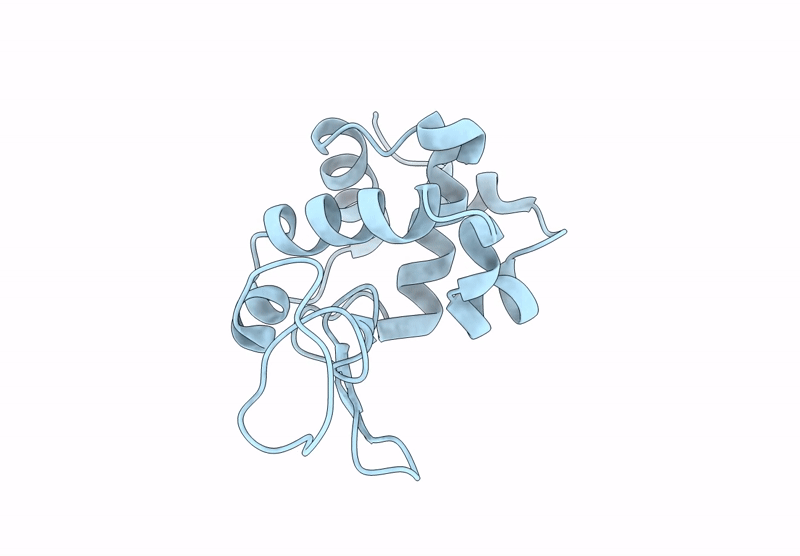
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Room-Temperature Structure Of Lysozyme Determined By Serial Synchrotron Crystallography
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: CL |
 |
Room Temperature Structure Of Lysozyme By Serial Synchrotron Crystallography (Xds)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-12 Classification: HYDROLASE |
 |
Room Temperature Structure Of Lysozyme By Serial Synchrotron Crystallography (Mosflm)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-12 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
 |
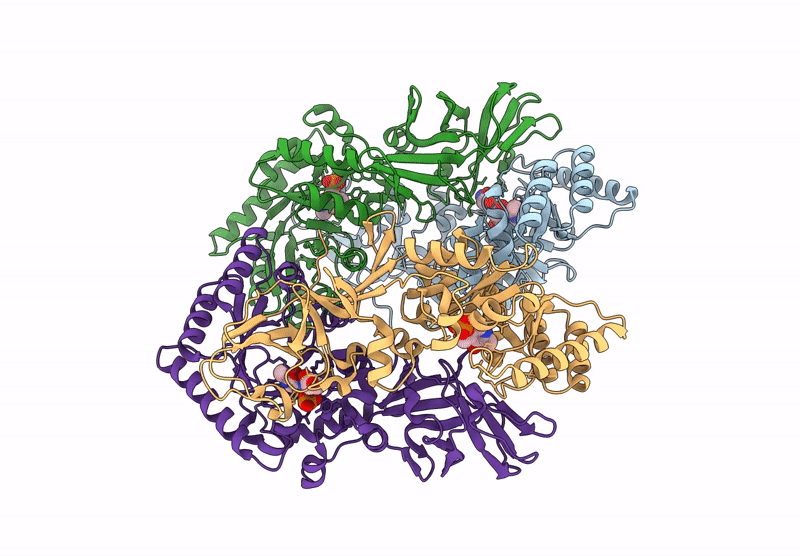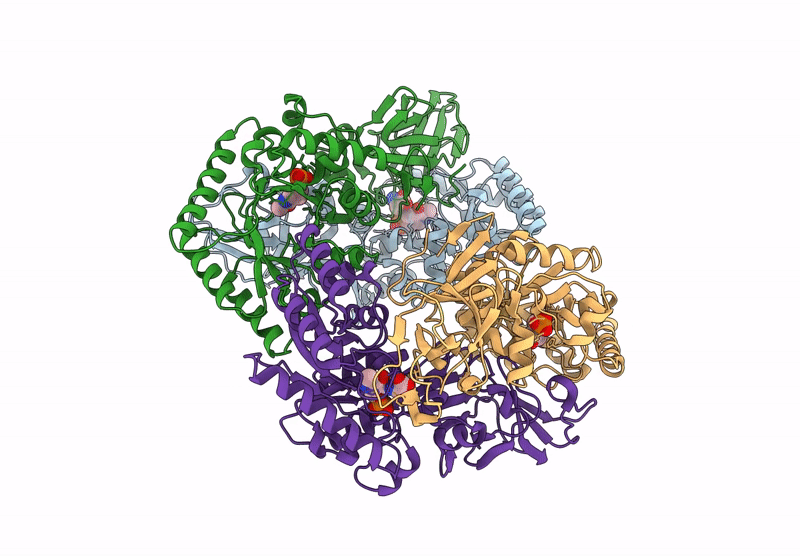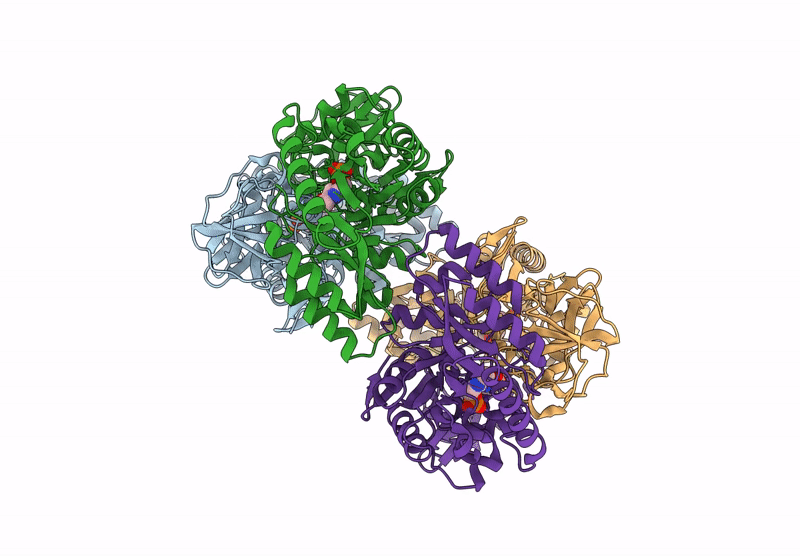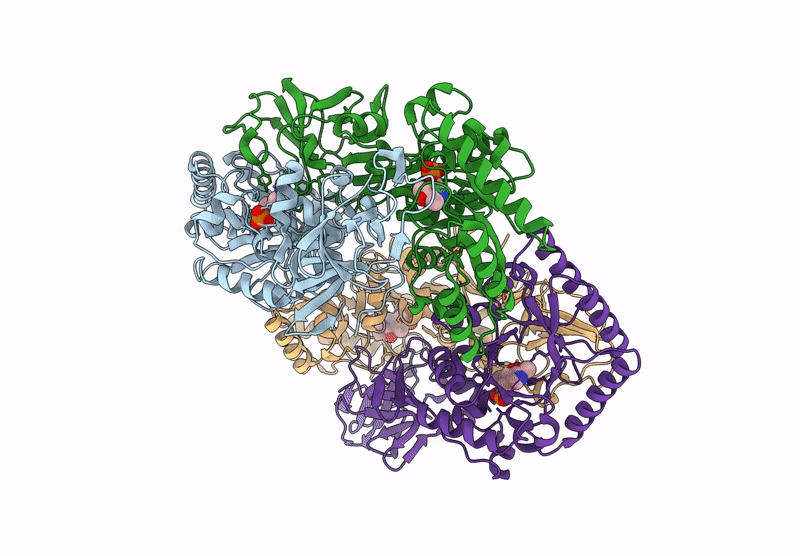
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, ALA, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
 |
Organism: Thermoanaerobacterium saccharolyticum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: TRS, NA |
 |
Organism: Thermoanaerobacterium saccharolyticum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: TRS, NA |
 |
Room-Temperature Structure Of Lysozyme Determined By Serial Synchrotron Crystallography (Mosflm)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-06 Classification: HYDROLASE |
 |
Room-Temperature Structure Of Lysozyme Determined By Serial Synchrotron Crystallography (Xgandalf)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-06 Classification: HYDROLASE |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-06-05 Classification: HYDROLASE Ligands: ZN, CA, ILE, LYS |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-06-05 Classification: HYDROLASE Ligands: ILE, LYS, ZN, CA |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-06-05 Classification: HYDROLASE Ligands: ILE, LYS, ZN, CA |
 |
Organism: Montipora sp. 20
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-04-17 Classification: FLUORESCENT PROTEIN |
 |
Organism: Thermoanaerobacterium saccharolyticum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-04-17 Classification: HYDROLASE Ligands: XYP, ACT |
 |
Organism: Thermoanaerobacterium saccharolyticum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-17 Classification: HYDROLASE Ligands: ACT, XYP |

