Search Count: 299
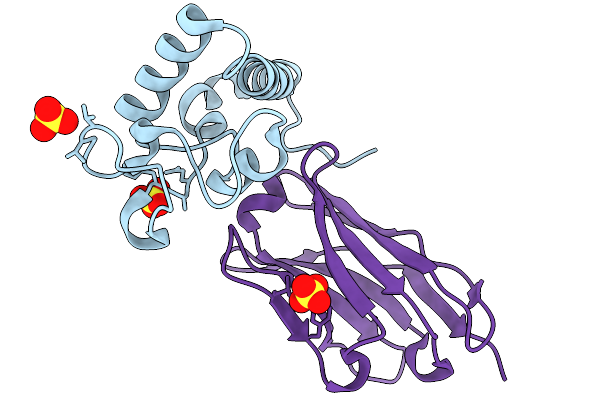 |
Crystal Structure Of Tetraspanin Cd63Mutant Large Extracellular Loop (Lel) In Complex With Sybody Sb3
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: CELL ADHESION Ligands: SO4 |
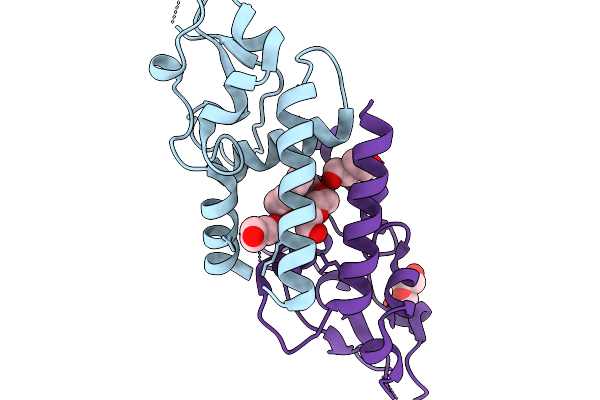 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: ENDOCYTOSIS Ligands: PEG |
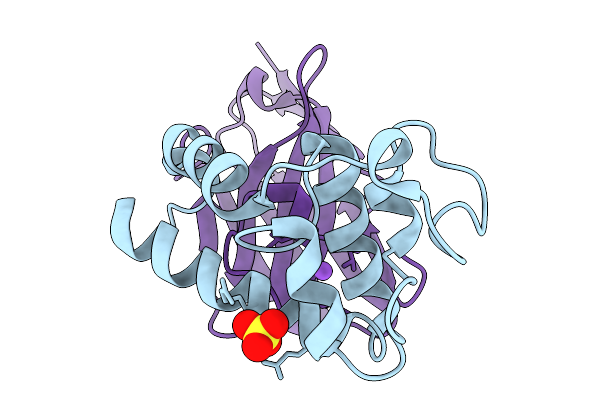 |
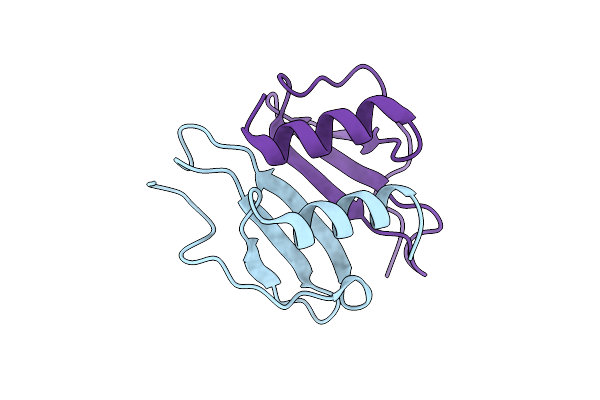
Crystal Structure Of Tetraspanin Cd63Mutant Large Extracellular Loop (Lel) In Complex With Sybody La4
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: CELL ADHESION Ligands: SO4, NA |
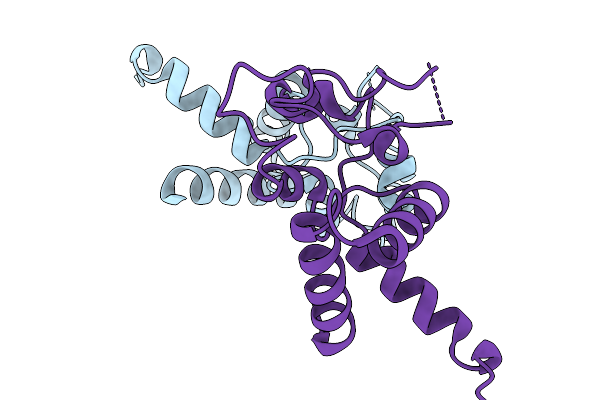 |
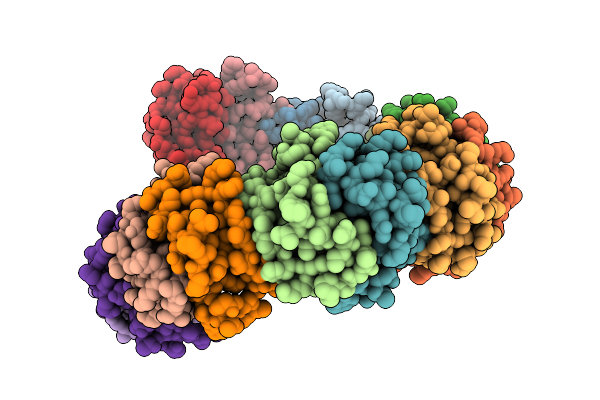
Crystal Structure Of Tetraspanin Tspan10Mutant Large Extracellular Loop (Lel)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: SIGNALING PROTEIN |
 |
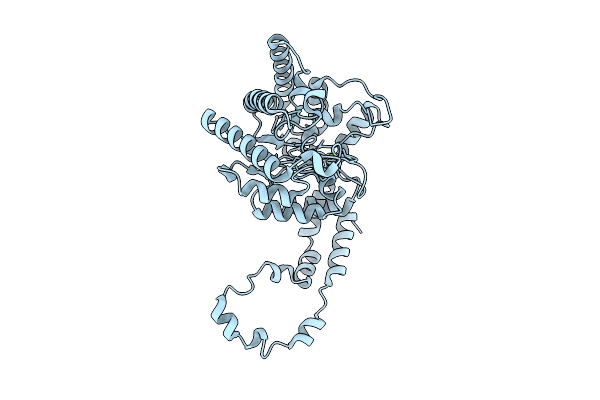
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: CYTOKINE Ligands: NO3 |
 |
 |
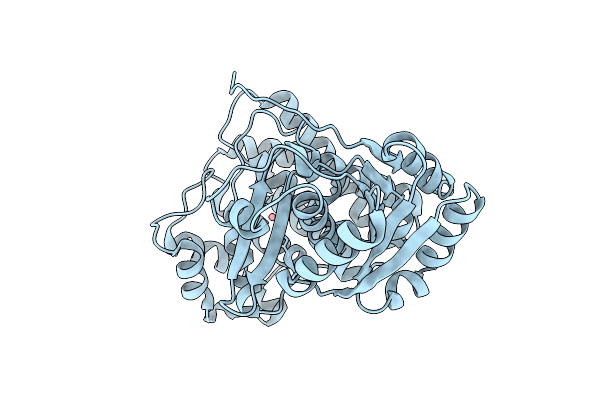
Serial Femtosecond Crystallography Structure Of Myoglobin From Equine Skeletal Muscle
Organism: Equus caballus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXYGEN STORAGE Ligands: SO4, HEM |
 |
Room Temperature Structure Of Glucose Isomerase By Serial Femtosecond Crystallography
Organism: Streptomyces pseudogriseolus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: ISOMERASE Ligands: MG |
 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: BIOSYNTHETIC PROTEIN Ligands: CO |
 |
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: BIOSYNTHETIC PROTEIN Ligands: CO, ADP |
 |
Organism: Thermoascus crustaceus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE |
 |
Organism: Cellulomonas biazotea
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Beta-Glucosidase Cba3 From Cellulomonas Biazotea In Complex With Glucose
Organism: Cellulomonas biazotea
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: BGC |
 |
Organism: Montipora sp. 20
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-23 Classification: FLUORESCENT PROTEIN |
 |
Room Temperature Structure Of Lysozyme By Serial Synchrotron Crystallography
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-03-05 Classification: STRUCTURAL PROTEIN Ligands: UI8, EDO, PLM, STE |
 |
Room-Temperature Structure Of Lysozyme Determined By Serial Synchrotron Crystallography
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: CL |
 |
Room Temperature Structure Of Lysozyme By Serial Synchrotron Crystallography (Xds)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-12 Classification: HYDROLASE |
 |
Room Temperature Structure Of Lysozyme By Serial Synchrotron Crystallography (Mosflm)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-12 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: CL |

