Search Count: 14
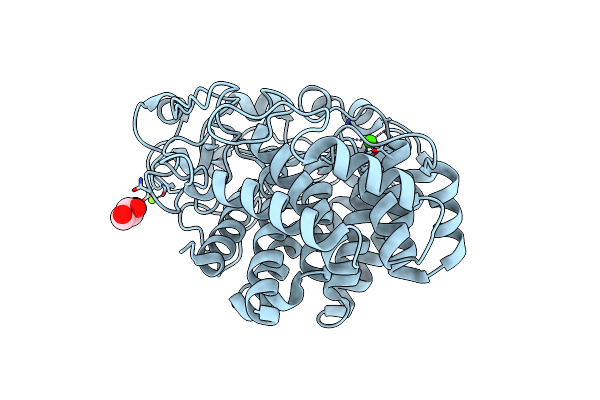 |
X-Ray Crystal Structure Of D43R Mutant Of Endo-1,4-Beta Glucanase From Eisenia Fetida
Organism: Eisenia fetida
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-12-06 Classification: HYDROLASE Ligands: GOL, MG, CA |
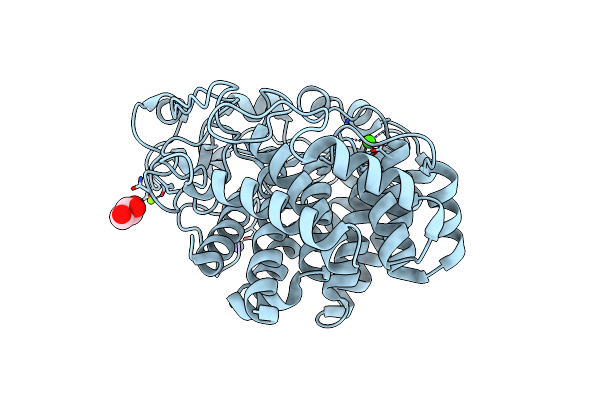 |
X-Ray Crystal Structure Of N372D Mutant Of Endo-1,4-Beta Glucanase From Eisenia Fetida
Organism: Eisenia fetida
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-12-06 Classification: HYDROLASE Ligands: MG, CA, NA, GOL |
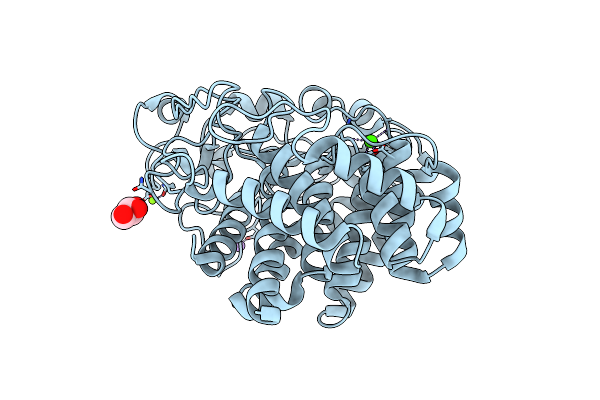 |
X-Ray Crystal Structure Of Q387E Mutant Of Endo-1,4-Beta Glucanase From Eisenia Fetida
Organism: Eisenia fetida
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-12-06 Classification: HYDROLASE Ligands: MG, CA, NA, GOL |
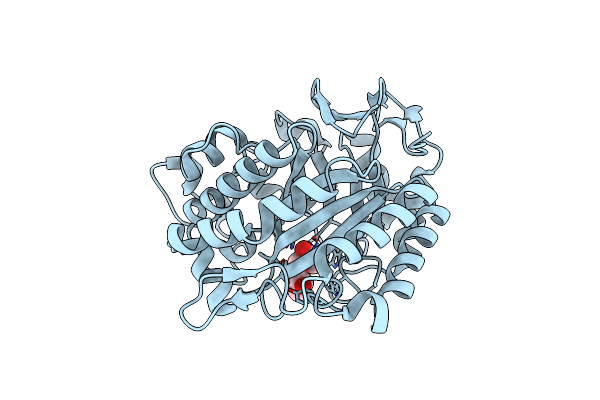 |
Organism: Eisenia fetida
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-06-27 Classification: CARBOHYDRATE Ligands: TRS, IPA |
 |
Crystal Structure Of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed With Unsaturated Galacturonosyl Rhamnose Substituted With Galactose
Organism: Penicillium chrysogenum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-04-04 Classification: LYASE Ligands: CA |
 |
Crystal Structure Of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum
Organism: Penicillium chrysogenum
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2018-03-21 Classification: LYASE Ligands: CA |
 |
Crystal Structure Of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed With Unsaturated Galacturonosyl Rhamnose
Organism: Penicillium chrysogenum
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2018-03-21 Classification: LYASE Ligands: CA |
 |
Crystal Structure Of A Pl 26 Exo-Rhamnogalacturonan Lyase From Penicillium Chrysogenum Complexed With Tetrameric Substrate
Organism: Penicillium chrysogenum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-03-21 Classification: LYASE Ligands: CA |
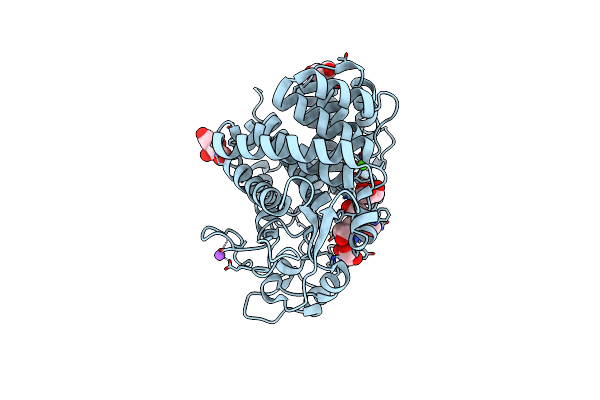 |
Organism: Eisenia fetida
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-10-30 Classification: HYDROLASE Ligands: CA, NA, TRS, FLC, GOL |
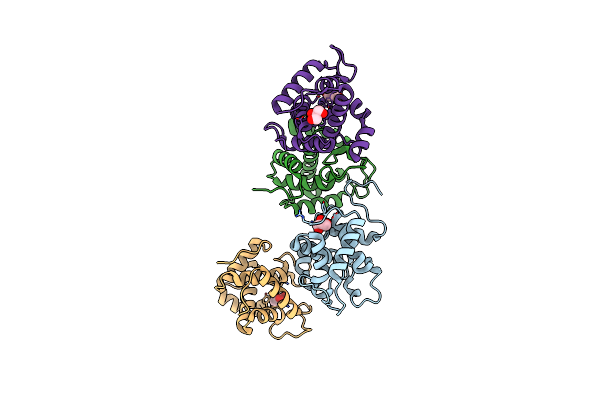 |
Crystal Structure Of Catalytic Domain Of Chitinase From Ralstonia Sp. A-471
Organism: Ralstonia
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: GOL |
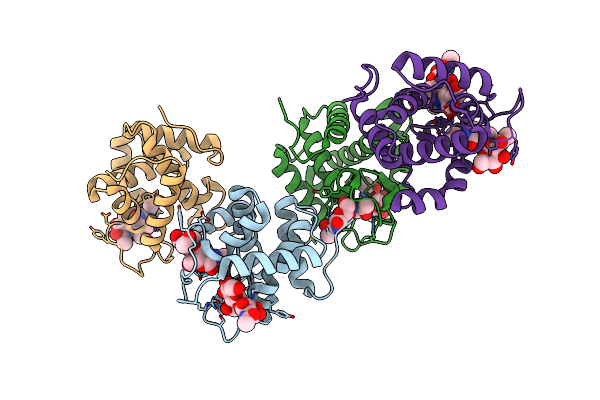 |
Crystal Structure Of Catalytic Domain Of Chitinase From Ralstonia Sp. A-471 In Complex With Disaccharide
Organism: Ralstonia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-05-15 Classification: HYDROLASE |
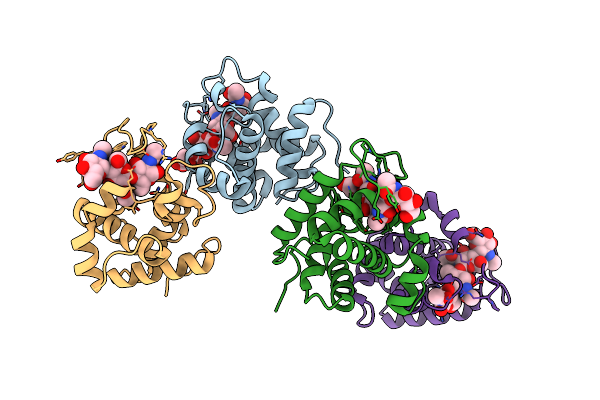 |
Crystal Structure Of Catalytic Domain Of Chitinase From Ralstonia Sp. A-471 (E141Q) In Complex With Tetrasaccharide
Organism: Ralstonia
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-05-15 Classification: HYDROLASE |
 |
Crystal Structure Of Catalytic Domain Of Chitinase From Ralstonia Sp. A-471 (E162Q)
Organism: Ralstonia
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: EPE, TRS |
 |
Crystal Structure Of Catalytic Domain Of Chitinase From Ralstonia Sp. A-471 (E162Q) In Complex With Disaccharide
Organism: Ralstonia
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-05-15 Classification: HYDROLASE |

