Search Count: 76
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1 Complexed With F Protein Hr1 (Hr1-42) (H32 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: MG |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1 Complexed With F Protein Hr1 (Hr1-42) (P21212 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: MG |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1 Complexed With F Protein Hr1 (Hr1-47) (P321 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: CA |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1Ek Complexed With F Protein Hr1 (Hr1-42) (P21 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek28 Complexed With F Protein Hr1 (Hr1-40) (H3 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: MPD |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek35Ge Complexed With F Protein Hr1 (Hr1-42) (P21212 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: ZN, ACT |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek35Ge Complexed With F Protein Hr1 (Hr1-42) (P2 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek35Gt Complexed With F Protein Hr1 (Hr1-42) (P21 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Cyanidioschyzon merolae (strain 10d)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-02-02 Classification: TRANSPORT PROTEIN Ligands: ANP, ZN, MG, CL, ACT |
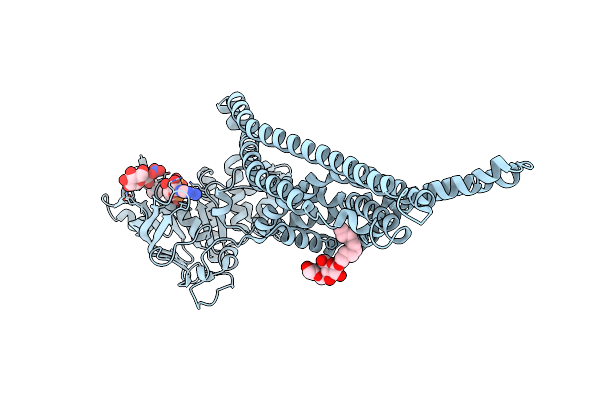 |
Organism: Cyanidioschyzon merolae (strain 10d)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-03-24 Classification: TRANSPORT PROTEIN Ligands: ANP, DMU, NO3, MG |
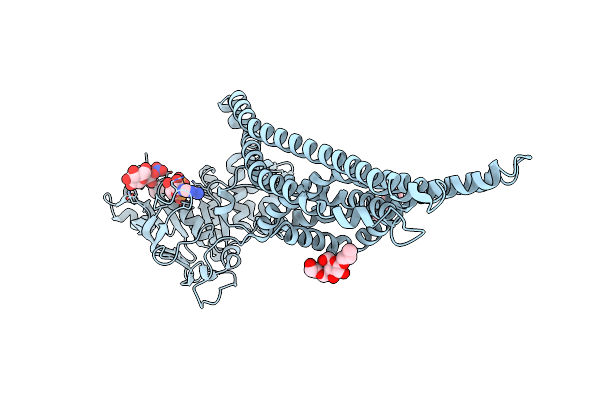 |
Crystal Structure Of An Outward-Open Nucleotide-Bound State Of The Eukaryotic Abc Multidrug Transporter Cmabcb1
Organism: Cyanidioschyzon merolae strain 10d
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-02-20 Classification: TRANSPORT PROTEIN Ligands: ANP, MG, DMU, NO3 |
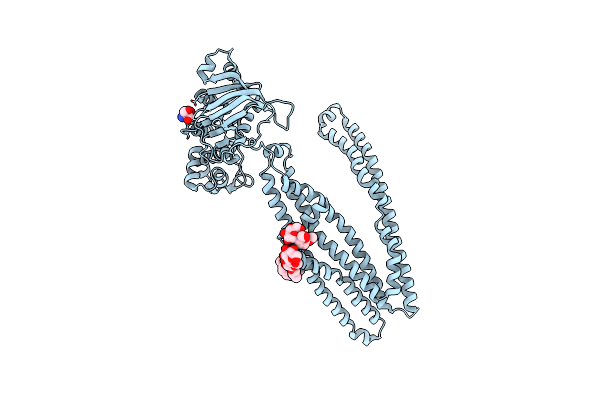 |
Crystal Structure Of An Inward-Open Apo State Of The Eukaryotic Abc Multidrug Transporter Cmabcb1
Organism: Cyanidioschyzon merolae strain 10d
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2019-01-16 Classification: TRANSPORT PROTEIN Ligands: DMU, TRS |
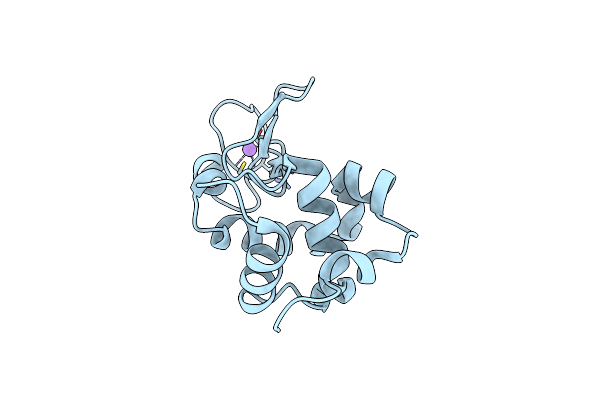 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2017-12-20 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-12-06 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-12-06 Classification: HYDROLASE |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-11-29 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Engyodontium album
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-11-29 Classification: HYDROLASE Ligands: NO3, PR |
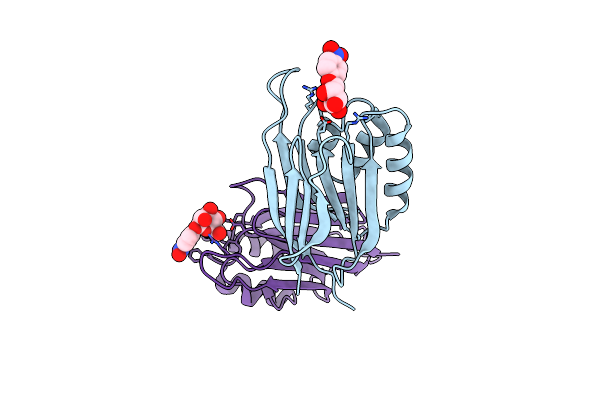 |
Serial Femtosecond X-Ray Structure Of A Stem Domain Of Human O-Mannose Beta-1,2-N-Acetylglucosaminyltransferase Solved By Se-Sad Using Xfel (Refined Against 13,000 Patterns)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-08-30 Classification: SUGAR BINDING PROTEIN Ligands: MBE |
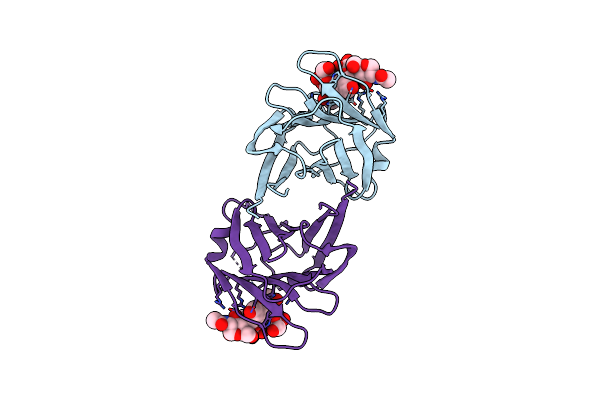 |
Serial Femtosecond X-Ray Structure Of Agrocybe Cylindracea Galectin With Lactose Solved By Se-Sad Using Xfel (Refined Against 60,000 Patterns)
Organism: Agrocybe cylindracea
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-08-30 Classification: SUGAR BINDING PROTEIN |
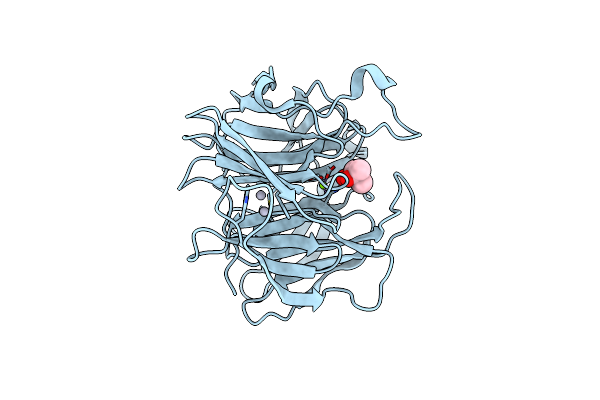 |
Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns)
Organism: Photinus pyralis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-08-30 Classification: HYDROLASE Ligands: HG, MG, MPD |

