Search Count: 47
 |
Organism: Takifugu rubripes
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-02 Classification: IMMUNE SYSTEM Ligands: GLC, BGC, CA |
 |
Organism: Takifugu rubripes
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-04-02 Classification: IMMUNE SYSTEM Ligands: GOL, CA |
 |
Organism: Fusobacterium nucleatum subsp. nucleatum atcc 25586
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2019-07-10 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
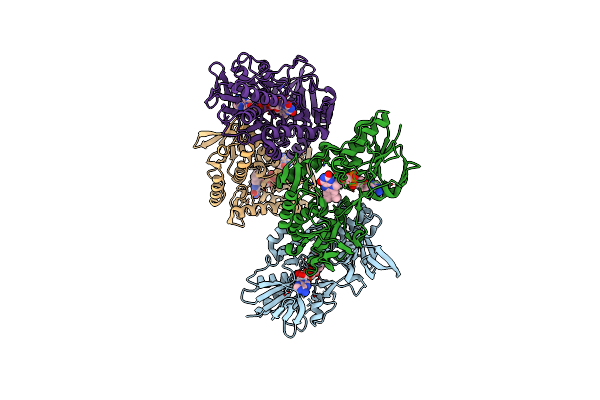
Structure Of Bacterial Type Ii Nadh Dehydrogenase From Caldalkalibacillus Thermarum Complexed With A Quinone Inhibitor Hqno At 2.8A Resolution
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: FAD, HQO |
 |
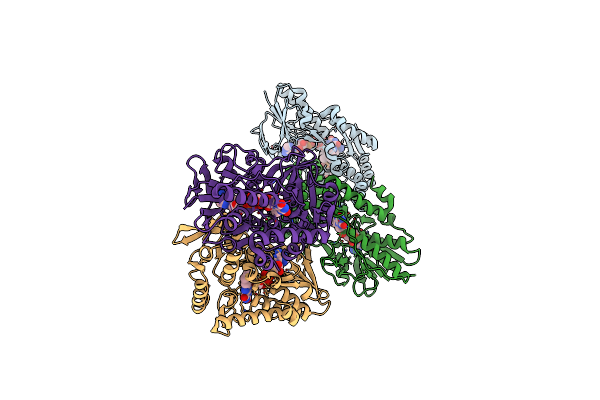
Structure Of Bacterial Type Ii Nadh Dehydrogenase From Caldalkalibacillus Thermarum At 2.15A Resolution
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-10-18 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
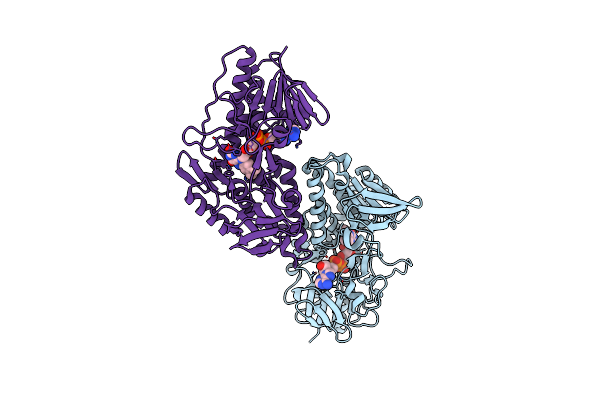
The Structure Of Type Ii Nadh Dehydrogenase From Caldalkalibacillus Thermarum Complexed With Nad+ At 2.5 Angstrom Resolution.
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-02-01 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
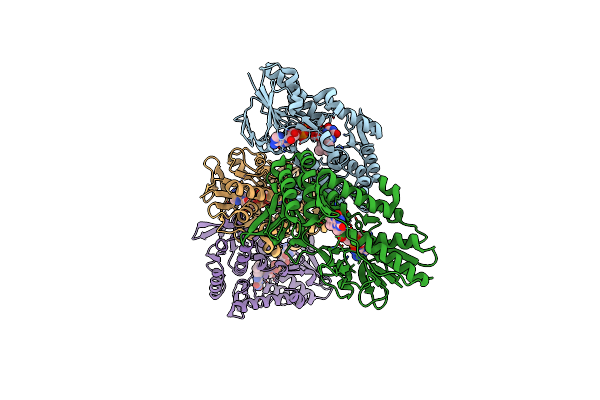
The Structure Of G164E Variant Of Type Ii Nadh Dehydrogenase From Caldalkalibacillus Thermarum
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2017-01-25 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
The Structure Of I379E Variant Of Type Ii Nadh Dehydrogenase From Caldalkalibacillus Thermarum
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-01-25 Classification: OXIDOREDUCTASE Ligands: FAD |
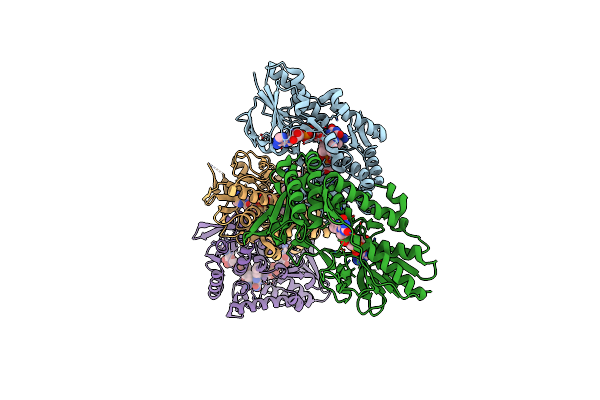 |
The Structure Of Type Ii Nadh Dehydrogenase From Caldalkalibacillus Thermarum Complexed With Nad+ At 3.0 Angstrom Resolution.
Organism: Caldalkalibacillus thermarum ta2.a1
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-01-25 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
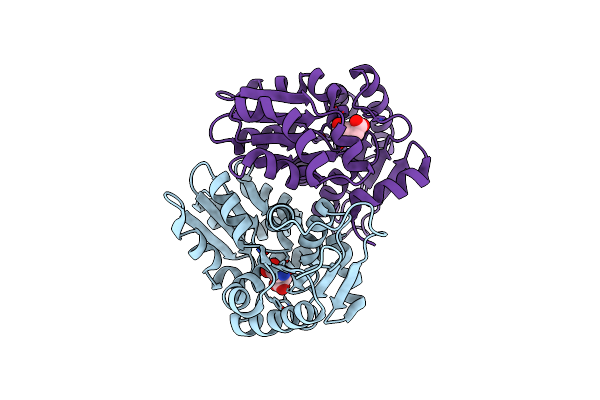 |
Glutamate Racemase Mycobacterium Tuberculosis (Muri) With Bound D-Glutamate, 2.3 Angstrom Resolution, X-Ray Diffraction
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-05-25 Classification: ISOMERASE Ligands: DGL |
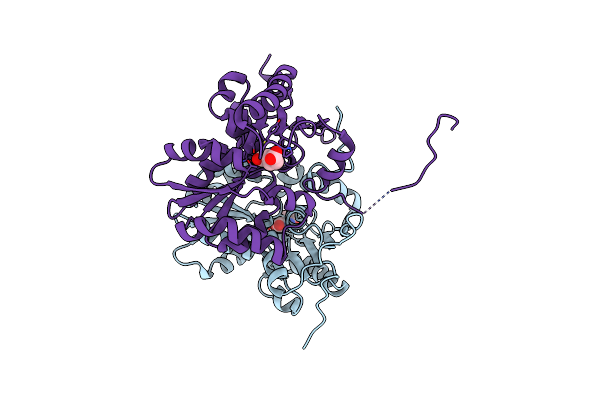 |
Glutamate Racemase (Muri) From Mycobacterium Smegmatis With Bound D-Glutamate, 1.8 Angstrom Resolution, X-Ray Diffraction
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-05-25 Classification: ISOMERASE Ligands: DGL, IOD |
 |
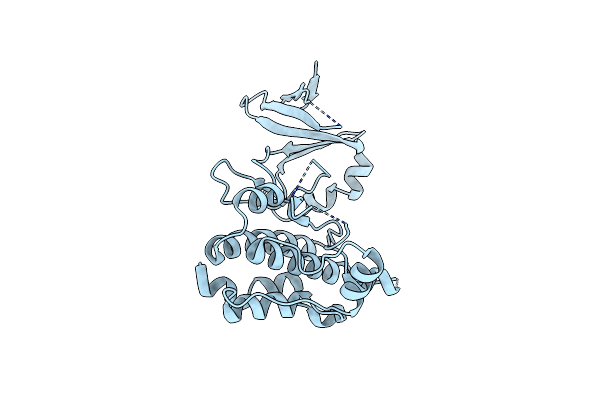
Organism: Pectobacterium atrosepticum (strain scri 1043 / atcc baa-672)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-03-16 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-11-11 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-11-11 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Orf virus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-07-15 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Orf virus (strain nz2), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-07-08 Classification: Viral Protein/cytokine |
 |
Organism: Orf virus (strain nz2), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-07-08 Classification: Viral Protein/cytokine |
 |
Organism: Orf virus (strain nz2), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2015-07-08 Classification: Viral Protein/cytokine |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-05-06 Classification: ISOMERASE |
 |
The Structure Of E292S Glycosynthase Variant Of Exo-1,3-Beta-Glucanase From Candida Albicans At 1.85A Resolution
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2014-06-25 Classification: HYDROLASE |

