Search Count: 30
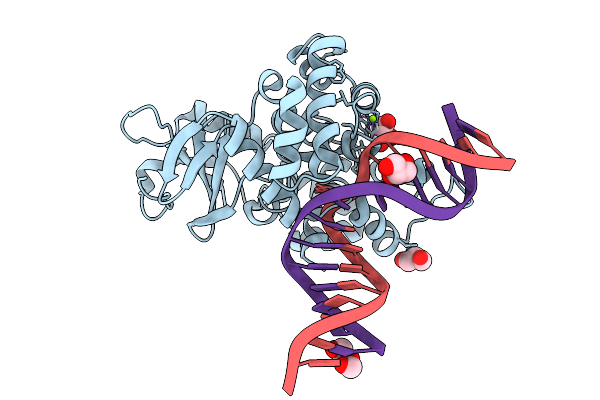 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Substrate 8-Oxoguanine Dna At Ph 8.0 Under 277 K
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: PEG, GOL, MG, NA |
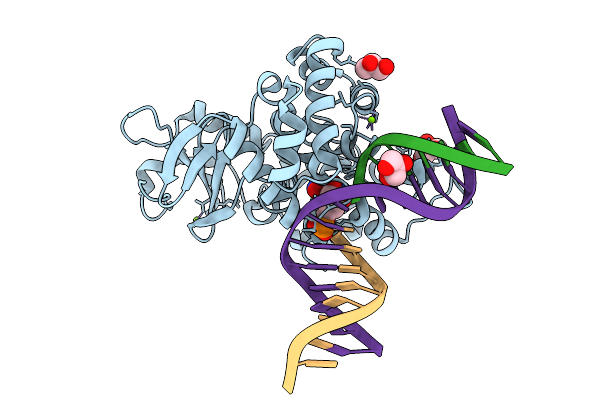 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 24 Hourss
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
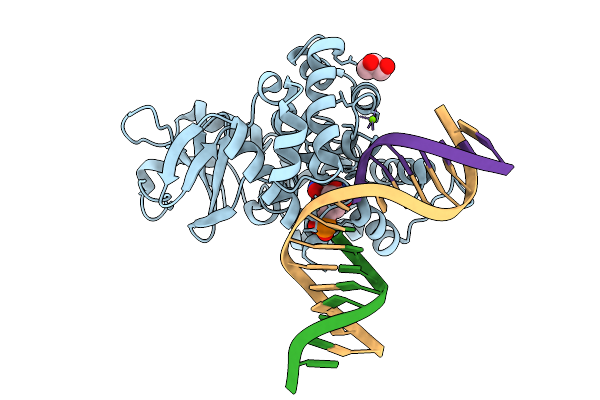 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 2.5 Hours
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
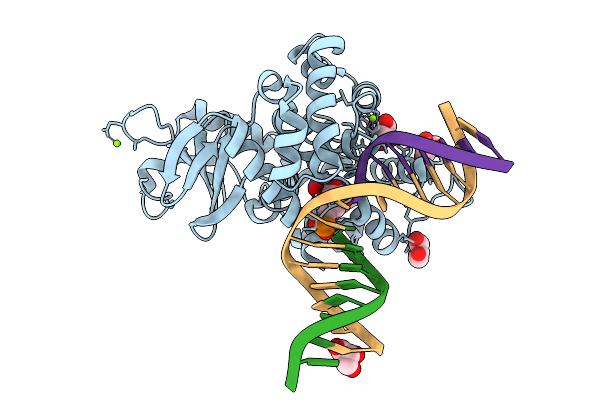 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 298 K For 3 Weeks
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, GOL, MG, NA |
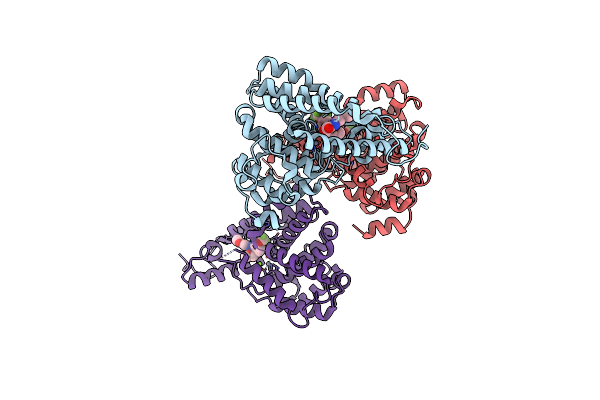 |
Discovery Of Clinical Candidate N-{(1S)-1-[3-Fluoro-4-(Trifluoromethoxy)Phenyl]-2-Methoxyethyl}-7-Methoxy-2-Oxo-2,3-Dihydropyrido[2,3-B]Pyrazine-4(1H)-Carboxamide (Tak-915), A Highly Potent, Selective, And Brain-Penetrating Phosphodiesterase 2A Inhibitor For The Treatment Of Cognitive Disorders
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, MG, 9GJ |
 |
Crystal Structure Of Human Phosphodiesterase 2A In Complex With 6-Methyl-N-(1-(4-(Trifluoromethoxy)Phenyl)Propyl)Pyrazolo[1,5-A]Pyrimidine-3-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: MG, ZN, 87R |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2016-08-31 Classification: DNA Ligands: AG |
 |
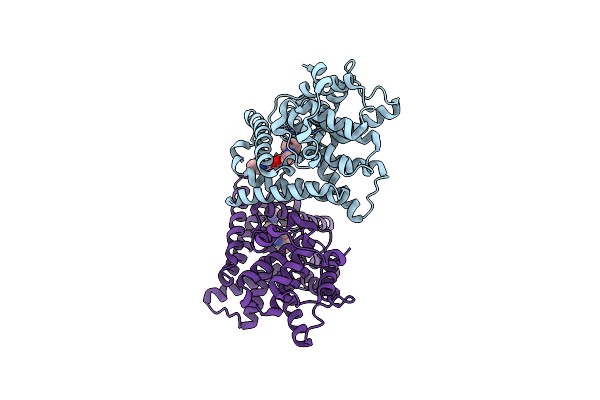
Crystal Structure Of The Catalytic Domain Of Human Pde10A Complexed With N-(4-((5-Methyl-5H-Pyrrolo[3,2-D]Pyrimidin-4-Yl)Oxy)Phenyl)-1H-Benzimidazol-2-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-06-29 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MG, ZN, 6DT |
 |
Crystal Structure Of The Catalytic Domain Of Human Pde10A Complexed With 1-(Cyclopropylmethyl)-5-(2-(2,3-Dihydro-1H-Imidazo[1,2-A]Benzimidazol-1-Yl)Ethoxy)-3-(1-Phenyl-1H-Pyrazol-5-Yl)Pyridazin-4(1H)-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-06-29 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MG, ZN, 6DW |
 |
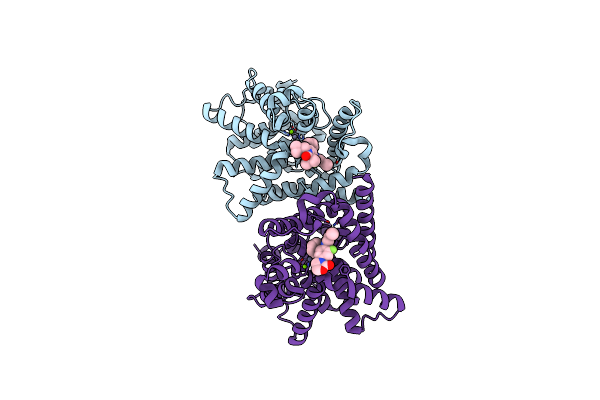
Crystal Structure Of The Catalytic Domain Of Pde10A Complexed With 1-(2-Fluoro-4-(2-Oxo-1,3-Oxazolidin-3-Yl)Phenyl)-5-Methoxy-3-(1-Phenyl-1H-Pyrazol-5-Yl)Pyridazin-4(1H)-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-11-11 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MG, ZN, 4LK |
 |
Crystal Structure Of The Catalytic Domain Of Pde10A Complexed With Highly Potent And Brain-Penetrant Pde10A Inhibitor With 2-Oxindole Scaffold
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2015-11-11 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MG, ZN, 4LP, 4LK |
 |
Crystal Structure Of The Catalytic Domain Of Pde10A Complexed With 3-(1-Phenyl-1H-Pyrazol-5-Yl)-1-(3-(Trifluoromethyl)Phenyl)Pyridazin-4(1H)-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-11-19 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MG, ZN, 3KG |
 |
Crystal Structure Of The Catalytic Domain Of Pde10A Complexed With 5-Methoxy-3-(1-Phenyl-1H-Pyrazol-5-Yl)-1-(3-(Trifluoromethyl)Phenyl)Pyridazin-4(1H)-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2014-11-19 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, MG, 3KB |
 |
Crystal Structure Of The Catalytic Domain Of Pde10A Complexed With 1-(2-Fluoro-4-(1H-Pyrazol-1-Yl)Phenyl)-5-Methoxy-3-(1-Phenyl-1H-Pyrazol-5-Yl)Pyridazin-4(1H)-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-11-19 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MG, ZN, 3K9 |
 |
Closed Conformation Of D-3-Hydroxybutyrate Dehydrogenase Complexed With Nad+ And L-3-Hydroxybutyrate
Organism: Pseudomonas fragi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-08-25 Classification: OXIDOREDUCTASE Ligands: NAD, 3HL, MG, GOL |
 |
Organism: Pseudomonas fragi
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-08-25 Classification: OXIDOREDUCTASE Ligands: NAD, 3HL, MG |
 |
Organism: Pseudomonas fragi
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-08-25 Classification: OXIDOREDUCTASE Ligands: NAD, MG |
 |
Organism: Pseudomonas fragi
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-08-25 Classification: OXIDOREDUCTASE Ligands: NAD, MG, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2006-08-01 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-08-01 Classification: HYDROLASE Ligands: ZN, SO4, BES |

