Search Count: 25
 |
Cryo-Em Structure Of Arabidopsis Thaliana Met1 (Aa:621-1534) In Complex With Hemimethylated Dna Analog
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: STRUCTURAL PROTEIN/DNA Ligands: SAH, ZN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: STRUCTURAL PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: STRUCTURAL PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-07-31 Classification: GENE REGULATION Ligands: ZN |
 |
Cryo-Em Structure Of Cdca7 Bound To Nucleosome Including Hemimethylated Cpg Site In Widom601 Positioning Sequence.
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2022-12-07 Classification: TRANSCRIPTION Ligands: ZN |
 |
Cryo-Em Structure Of Human Dnmt1 (Aa:351-1616) In Complex With Ubiquitinated H3 And Hemimethylated Dna Analog (Cxxc-Ordered Form)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: TRANSFERASE/DNA Ligands: ZN, SAH |
 |
Cryo-Em Structure Of Human Dnmt1 (Aa:351-1616) In Complex With Ubiquitinated H3 And Hemimethylated Dna Analog (Cxxc-Disordered Form)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: TRANSFERASE/DNA Ligands: ZN |
 |
Crystal Structure Of Human Uhrf1 Ttd In Complex With 5-Amino-2,4-Dimethylpyridine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-01-05 Classification: DNA BINDING PROTEIN Ligands: MPD, 8NF, DMS, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-10-09 Classification: LIGASE Ligands: ZN, EPE |
 |
Crystal Structure Of Dnmt1 Rfts Domain In Complex With K18/K23 Mono-Ubiquitylated Histone H3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-11-15 Classification: SIGNALING PROTEIN/TRANSFERASE Ligands: ZN |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2011-08-10 Classification: ISOMERASE Ligands: NDP, MN |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-08-10 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: NDP, MG, FOM, CA |
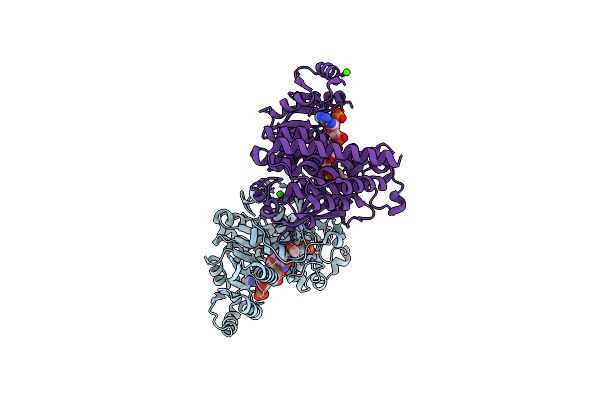 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2011-08-10 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: NDP, MG, F98, CA |
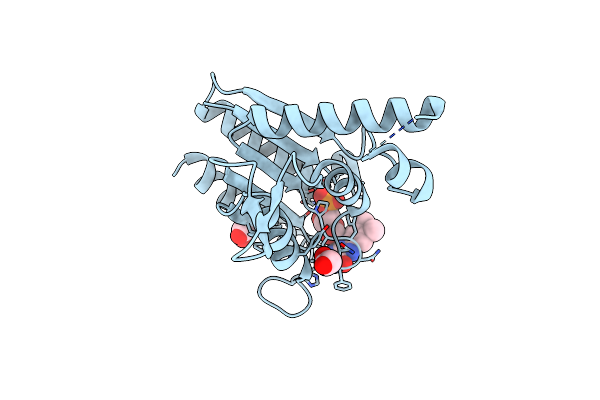 |
The Crystal Structure Of Azor (Azoreductase) From Escherichia Coli: Oxidized Azor In Tetragonal Crystals (The Resolution Has Improved From 1.8 (1V4B) To 1.4 Angstrom)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2008-03-11 Classification: OXIDOREDUCTASE Ligands: FMN, EDO |
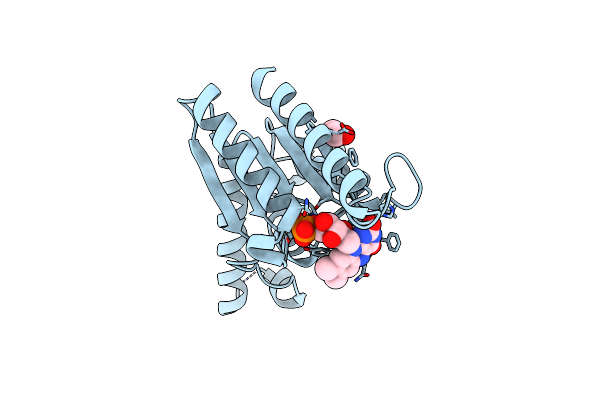 |
The Crystal Structure Of Azor (Azoreductase) From Escherichia Coli: Reduced Azor In Tetragonal Crystals
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-03-11 Classification: OXIDOREDUCTASE Ligands: FMN, EDO |
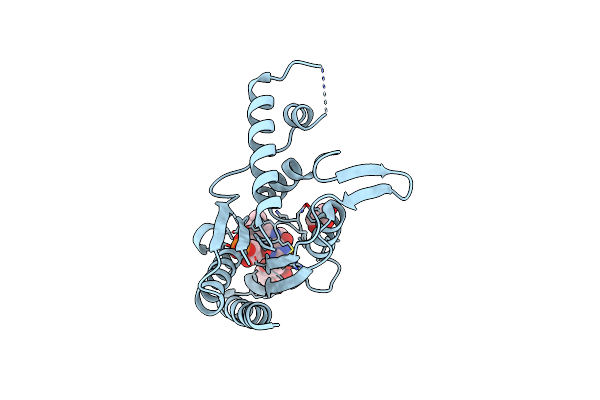 |
The Crystal Structure Of Azor (Azoreductase) From Escherichia Coli: Azor In Complex With Dicoumarol
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-03-11 Classification: OXIDOREDUCTASE Ligands: FMN, DTC, GOL |
 |
The Crystal Structure Of Azor (Azoreductase) From Escherichia Coli: Oxidized Azor In Orthorhombic Crystals
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-03-11 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-05-16 Classification: OXIDOREDUCTASE Ligands: FMN, GOL |

