Search Count: 166
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1S |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1R |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1S |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 100 Ns After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Dark State Sfx Structure Of Hen Egg White Lysozyme Microcrystals Immobilized With A Mn Carbonyl Complex At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 10 Ns After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 1 Microsecond After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 1 Microsecond After Photoexcitation With 40 Microjoules Laser Intensity At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Organism: Brevibacillus laterosporus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-07-10 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: EDO, SO4, CL |
 |
Cryo-Em Structure Of Groel Bound To Unfolded Substrate (Ugt1A) At 2.8 Ang. Resolution (Consensus Refinement)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Double Occupied Ring (Dor) Of Groel-Ugt1A Complex At 2.7 Ang. Resolution
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Single Empty Ring 2 (Ser2) Of Groel-Ugt1A Complex At 3.2 Ang. Resolution
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Occupied Ring Subunit 4 (Or4) Of Groel Complexed With Polyalanine Model Of Ugt1A From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Empty Ring Subunit 1 (Er1) From Single Empty Ring Of Groel-Ugt1A Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Empty Ring Subunit 2 (Er2) From Groel-Ugt1A Single Empty Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
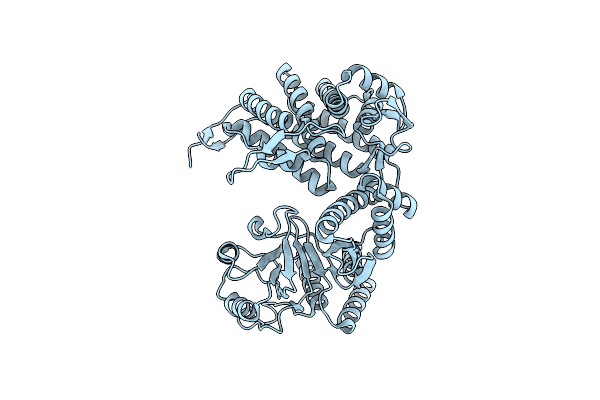 |
Cryo-Em Structure Of Occupied Ring Subunit 1 (Or1) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
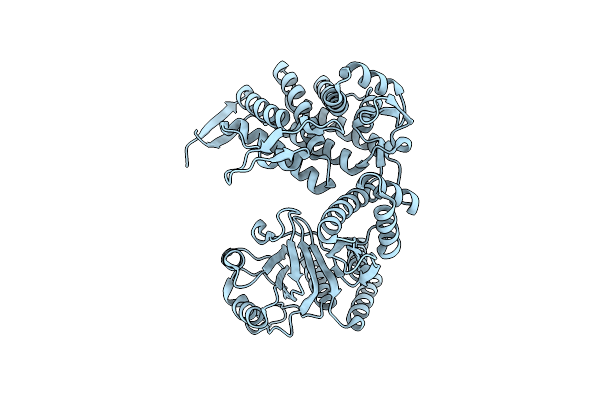 |
Cryo-Em Structure Of Occupied Ring Subunit 2 (Or2) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
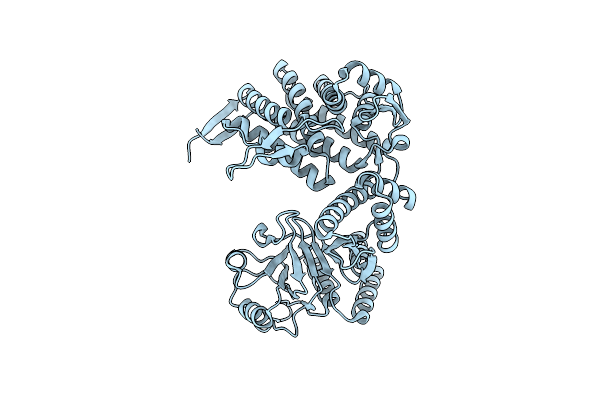 |
Cryo-Em Structure Of Occupied Ring Subunit 3 (Or3) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
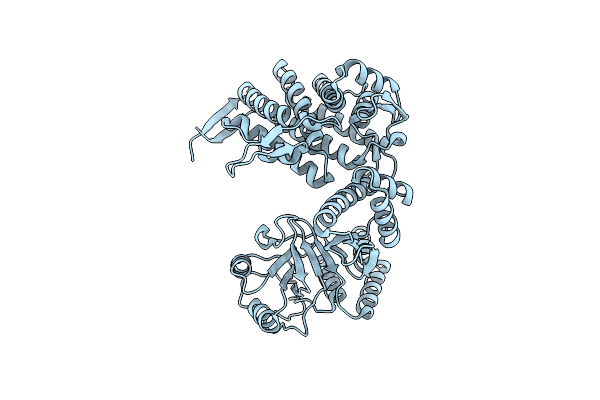 |
Cryo-Em Structure Of Occupied Ring Subunit 4 (Or4) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-03-02 Classification: METAL BINDING PROTEIN Ligands: RIR, EDO, SO4, CD, CL, IR |

