Search Count: 15
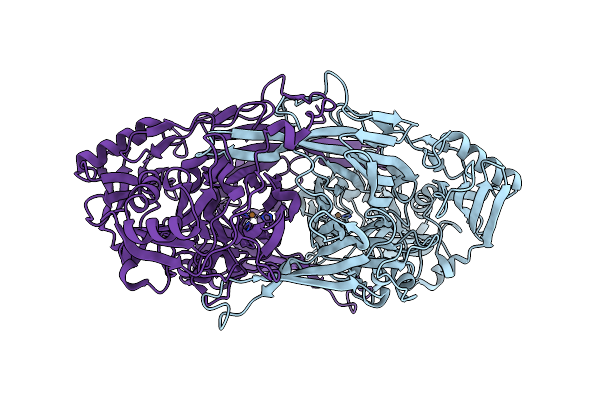 |
Crystal Structure Of Copper Amine Oxidase From Arthrobacter Globiformis: Substrate Schiff-Base Intermediate Formed With Ethylamine
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2008-08-05 Classification: OXIDOREDUCTASE Ligands: CU |
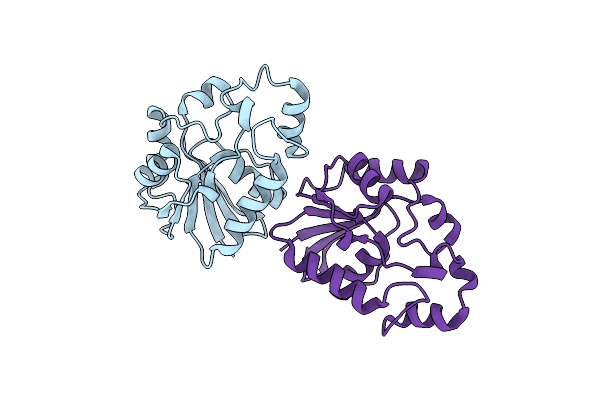 |
Structural Study Of Project Id Tthb049 From Thermus Thermophilus Hb8 (L52M)
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-10-02 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
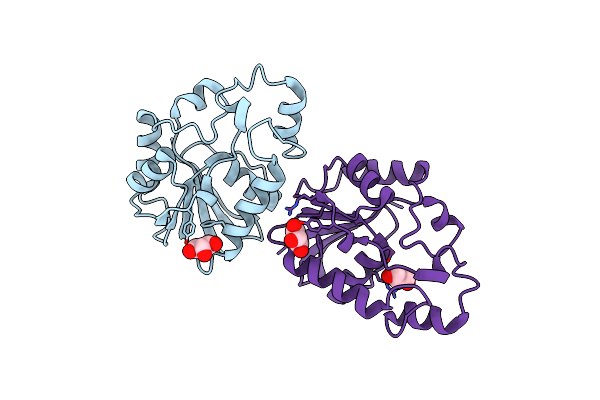 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: GOL |
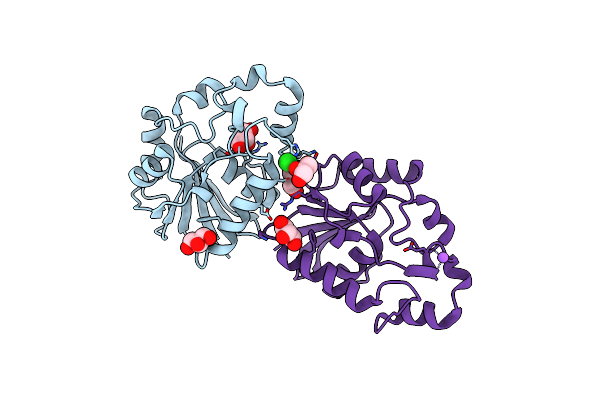 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: CL, GOL, NA |
 |
Structural Study Of Project Id Ph0725 From Pyrococcus Horikoshii Ot3 (E140M)
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-09-25 Classification: TRANSFERASE |
 |
Structural Study Of Project Id Ph0725 From Pyrococcus Horikoshii Ot3 (L202M)
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-09-25 Classification: TRANSFERASE |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-09-25 Classification: HYDROLASE Ligands: GOL, NA |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-08-21 Classification: TRANSFERASE Ligands: SO4, SAH, GOL |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-08-21 Classification: TRANSFERASE |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-08-21 Classification: TRANSFERASE Ligands: SAH, SO4 |
 |
Structural Study Of Project Id Ph0725 From Pyrococcus Horikoshii Ot3 (K19M)
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-07-24 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-07-24 Classification: TRANSFERASE |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-06-19 Classification: TRANSFERASE |
 |
Substrate Schiff-Base Intermediate With Tyramine In Copper Amine Oxidase From Arthrobacter Globiformis
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2006-05-02 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2006-01-24 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |

