Search Count: 19
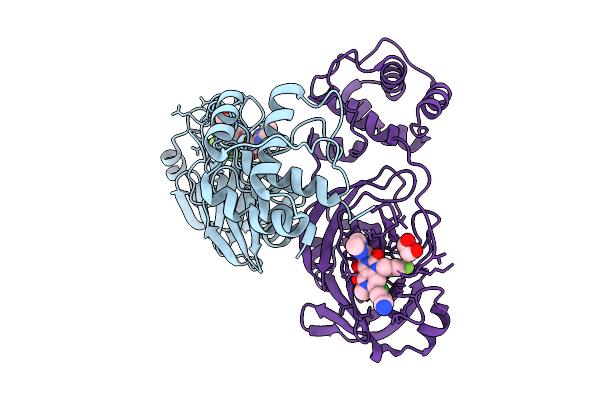 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: VIRAL PROTEIN Ligands: A1MA2, EDO |
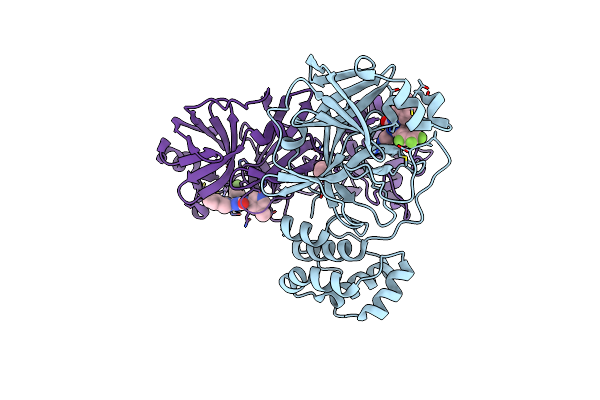 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: A1L7P, EDO |
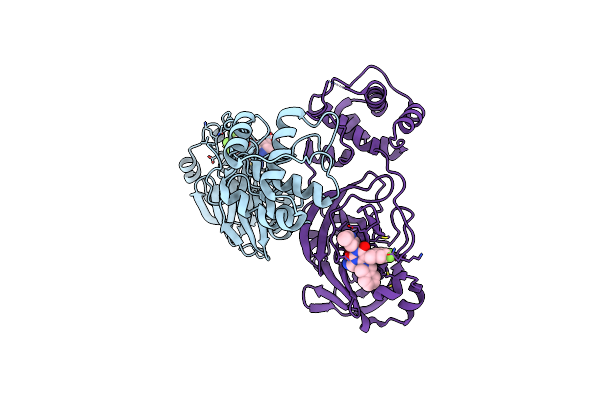 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: A1L7Q |
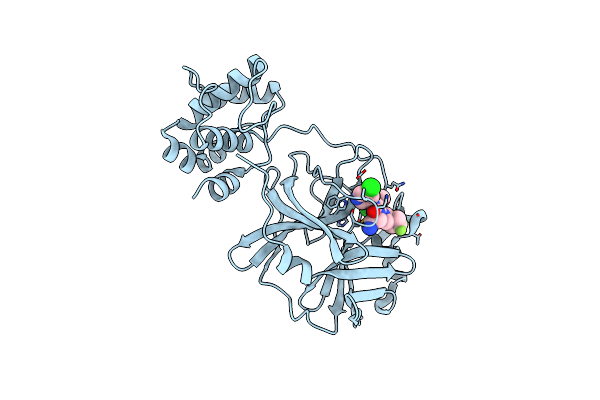 |
Crystal Structure Of Sars-Cov-2 3Cl Protease In Complex With Compound 17 (S-892216)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: A1L7R |
 |
Crystal Structure Of An Inverse Agonist Antipsychotic Drug Derivative-Bound 5-Ht2C
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2024-08-28 Classification: MEMBRANE PROTEIN Ligands: A1L10 |
 |
Crystal Structure Of An Inverse Agonist Antipsychotic Drug Pimavanserin-Bound 5-Ht2A
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2024-08-28 Classification: MEMBRANE PROTEIN Ligands: A1L11 |
 |
The Crystal Structure Of Sars-Cov-2 3Cl Protease In Complex With Compound 1
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-06 Classification: VIRAL PROTEIN/INHIBITOR Ligands: 7XB |
 |
The Crystal Structure Of Sars-Cov-2 3Cl Protease In Complex With Compound 3
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-04-06 Classification: Viral protein/inhibitor Ligands: 7YY |
 |
Crystal Structure Of Bace1 In Complex With N-{3-[(3S)-1-Amino-5-Fluoro-3-Methyl-3,4-Dihydro-2,6-Naphthyridin-3-Yl]-4-Fluorophenyl}-5-Cyano-3-Methylpyridine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-07-28 Classification: HYDROLASE Ligands: IOD, GOL, GUC |
 |
Crystal Structure Of Bace1 In Complex With N-{3-[(4S,6S)-2-Amino-4-Methyl-6-(Trifluoromethyl)-5,6-Dihydro-4H-1,3-Thiazin-4-Yl]-4-Fluorophenyl}-5-(Fluoromethoxy)Pyrazine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: IOD, GOL, C86 |
 |
Crystal Structure Of Bace1 In Complex With N-{3-[(4R,5R,6R)-2-Amino-5-Fluoro-4,6-Dimethyl-5,6-Dihydro-4H-1,3-Thiazin-4-Yl]-4-Fluorophenyl}-5-(Fluoromethoxy)Pyrazine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-10-30 Classification: HYDROLASE Ligands: IOD, GOL, DMS, C83 |
 |
Crystal Structure Of Bace1 In Complex With N-(3-((4S,5R)-2-Amino-4-Methyl-5-Phenyl-5,6-Dihydro-4H-1,3-Thiazin-4-Yl)-4-Fluorophenyl)-5-(Fluoromethoxy)Pyrazine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-08-28 Classification: HYDROLASE Ligands: IOD, DMS, C6R |
 |
Crystal Structure Of Bace1 In Complex With N-(3-((4S,5S)-2-Amino-4-Methyl-5-Phenyl-5,6-Dihydro-4H-1,3-Thiazin-4-Yl)-4-Fluorophenyl)-5-(Fluoromethoxy)Pyrazine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-08-28 Classification: HYDROLASE Ligands: IOD, DMS, C7X |
 |
Crystal Structure Of Bace1 In Complex With N-{3-[(4S)-2-Amino-4-Methyl-5,6-Dihydro-4H-1,3-Thiazin-4-Yl]-4-Fluorophenyl}-5-Chloropyridine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-08-28 Classification: HYDROLASE Ligands: IOD, NA, GOL, C6U |
 |
Crystal Structure Of Bace1 In Complex With N-{3-[(5R)-3-Amino-5-Methyl-9,9-Dioxo-2,9Lambda6-Dithia-4-Azaspiro[5.5]Undec-3-En-5-Yl]-4-Fluorophenyl}-5-(Fluoromethoxy)Pyrazine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-08-28 Classification: HYDROLASE Ligands: IOD, GOL, C7O |
 |
Bace2 Xaperone Complex With N-{3-[(5R)-3-Amino-5-Methyl-9,9-Dioxo-2,9Lambda6-Dithia-4-Azaspiro[5.5]Undec-3-En-5-Yl]-4-Fluorophenyl}-5-(Fluoromethoxy)Pyrazine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2019-08-28 Classification: HYDROLASE/IMMUNE SYSTEM Ligands: C7O, CL |
 |
Structure Of Bace1 In Complex With N-(3-((4R,5R,6S)-2-Amino-6-(1,1-Difluoroethyl)-5-Fluoro-4-Methyl-5,6-Dihydro-4H-1,3-Oxazin-4-Yl)-4-Fluorophenyl)-5-(Fluoromethoxy)Pyrazine-2-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-08-08 Classification: HYDROLASE/INHIBITOR Ligands: IOD, GOL, DMS, 0B5 |
 |
Crystal Structure Of Nitric Reductase From Denitrifying Fungus Fusarium Oxysporum
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1997-10-15 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Nitric Reductase From Denitrifying Fungus Fusarium Oxysporum Complex With Carbon Monoxide
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1997-10-15 Classification: OXIDOREDUCTASE Ligands: HEM, CMO |

