Search Count: 49
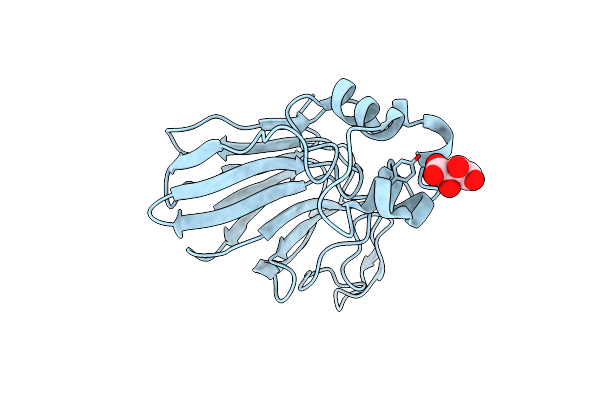 |
Magnetic Effects Of Thaumatin Crystals; Observation Of The Crystal Growth Of Magneto-Archimedes Levitation And Magnetic Orientation
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2025-02-26 Classification: PLANT PROTEIN Ligands: TLA |
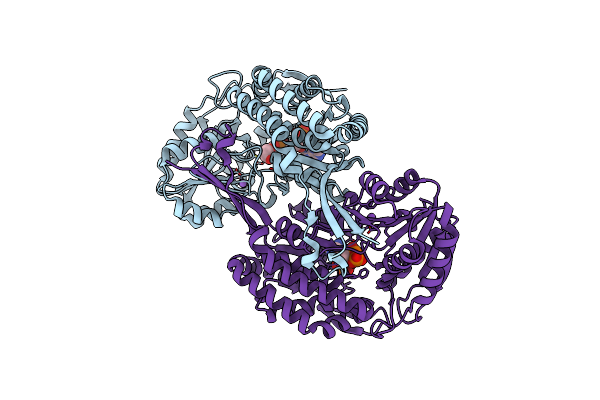 |
Crystal Structure Of Thermostable Acetaldehyde Dehydrogenase From Hyperthermophilic Archaeon Sulfolobus Tokodaii
Organism: Sulfurisphaera tokodaii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-06-07 Classification: OXIDOREDUCTASE Ligands: NA7, NA, ATR |
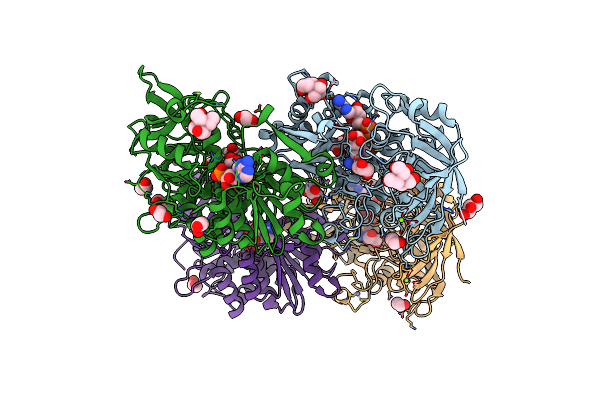 |
Organism: Linum usitatissimum
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-02-09 Classification: LYASE Ligands: NAD, ZN, GOL, MG, EDO, BTB |
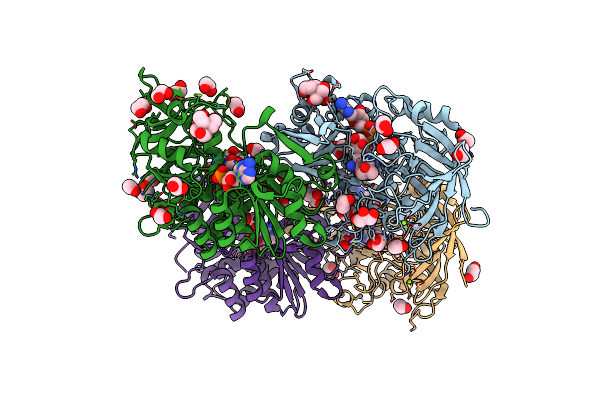 |
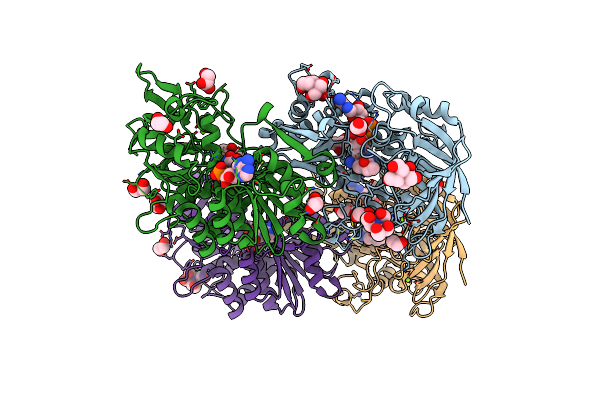
Crystal Structure Of Hydroxynitrile Lyase From Linum Usitatissimum Complexed With Acetone Cyanohydrin
Organism: Linum usitatissimum
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2022-02-09 Classification: LYASE Ligands: NAD, ZN, CNH, MG, EDO, GOL, BTB, ACN |
 |
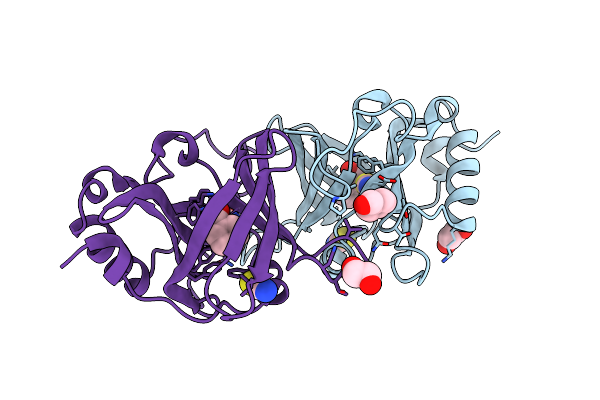
Crystal Structure Of Hydroxynitrile Lyase From Linum Usitatissium Complexed With (R)-2-Hydroxy-2-Methylbutanenitrile
Organism: Linum usitatissimum
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-02-09 Classification: LYASE Ligands: NAD, ZN, MG, 5Z9, EDO, GOL, BTB |
 |
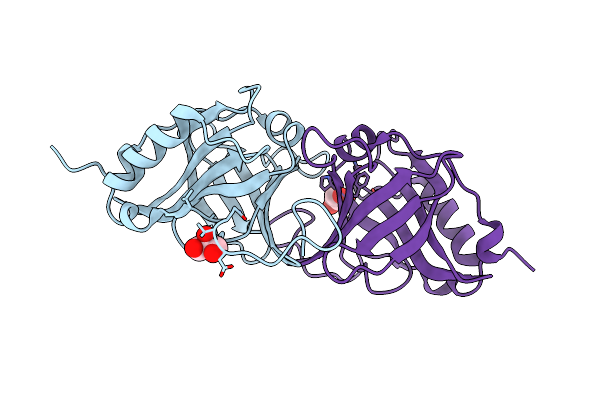
Hydroxynitrile Lyase From Parafontaria Laminate Complexed With Benzaldehyde Prepared By Cocrystallization
Organism: Parafontaria laminata
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2021-04-07 Classification: LYASE Ligands: HBX, SCN, EDO |
 |
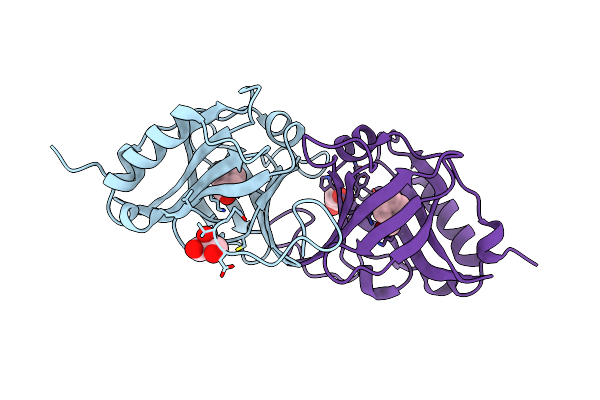
Organism: Parafontaria falcifera
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2021-03-24 Classification: LYASE Ligands: GOL |
 |
Hydroxynitrile Lyase From Parafonteria Laminate Complexed With Benzaldehyde
Organism: Parafontaria falcifera
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2021-03-24 Classification: LYASE Ligands: HBX, GOL |
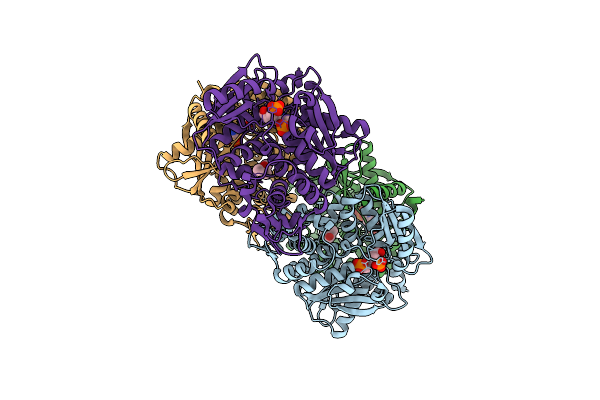 |
Crystal Structure Of The O-Phosphoserine Sulfhydrylase From Aeropyrum Pernix Complexed With O-Phosphoserine
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: E1U, MPD |
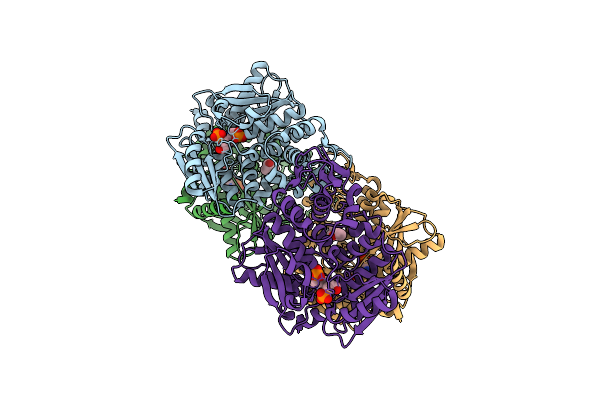 |
Crystal Structure Of The O-Phosphoserine Sulfhydrylase From Aeropyrum Pernix Complexed With O-Phosphoserine
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: E1U, MPD |
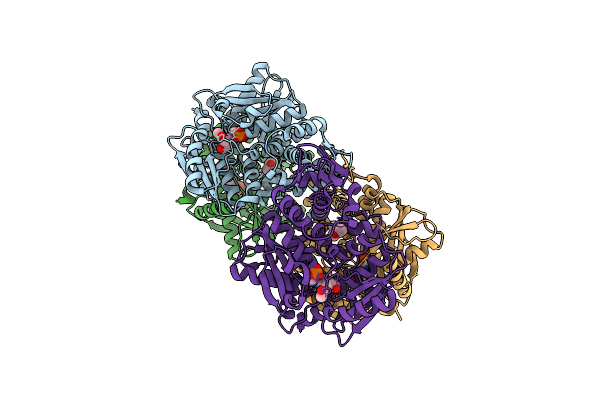 |
Crystal Structure Of The O-Phosphoserine Sulfhydrylase From Aeropyrum Pernix Complexed With O-Acetylserine
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: E1R, MPD |
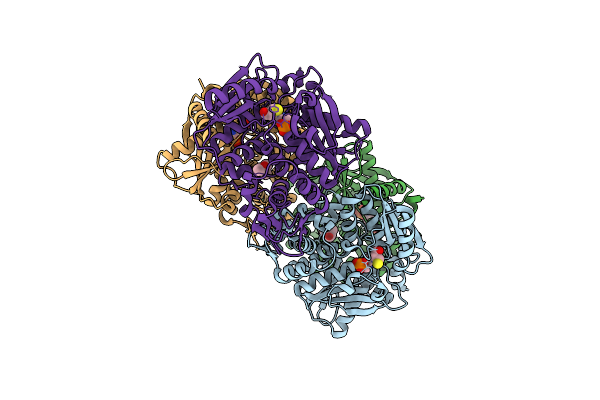 |
Crystal Structure Of The O-Phosphoserine Sulfhydrylase From Aeropyrum Pernix Complexed With L-Cysteine
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: E1O, MPD |
 |
 |
 |
 |
 |
Crystal Structure Of A Hypothetical Protein, Ttha0829 From Thermus Thermophilus Hb8, Composed Of Cystathionine-Beta-Synthase (Cbs) And Aspartate-Kinase Chorismate-Mutase Tyra (Act) Domains
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2016-05-18 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-11-18 Classification: TRANSCRIPTION Ligands: YSL |
 |
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-11-18 Classification: TRANSCRIPTION Ligands: YSE |
 |
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-04-29 Classification: HYDROLASE |

