Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
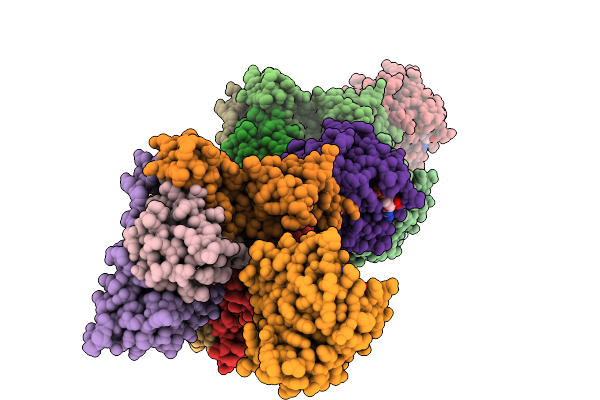
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME Ligands: MG, ZN, A1L8F |
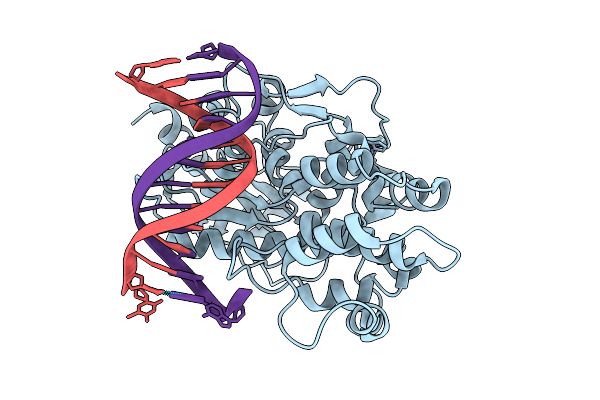 |
Crystal Structure Of Vanc21 In Complex With Its Target Dna (5-Bromouridine Substituted).
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Cryo-Em Structure Of Leminorella Grimontii Gatc In The Presence Of D-Xylose
Organism: Leminorella grimontii
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN |
 |
Organism: Leminorella grimontii
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN |
 |
Organism: Leminorella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN Ligands: OLC |
 |
X-Ray Structure Of Clostridioides Difficile Autolysin Acd33800 Catalytic Domain
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Cryo-Em Structure Of Chalcone Synthase (Chs) From Physcomitrella Patens In The Presence Of Chil
Organism: Physcomitrium patens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: TRANSFERASE |
 |
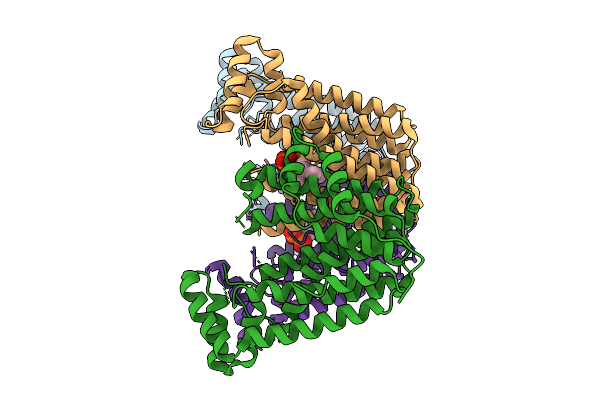
Organism: Streptomyces sp. ks84
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: SO4, EDO |
 |
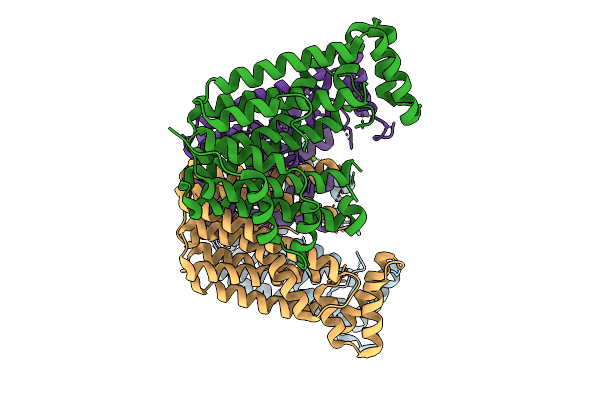
Organism: Streptomyces sp. ks84
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: FPS, MG |
 |
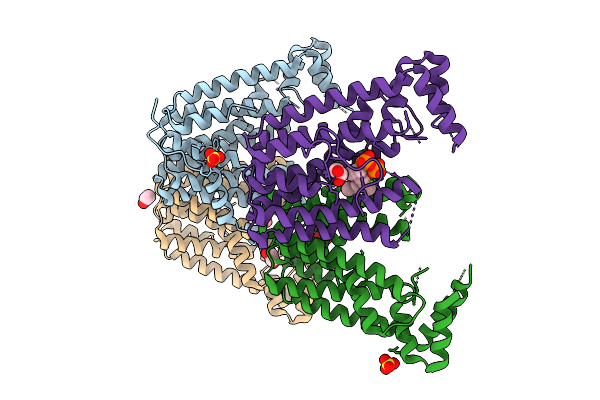
Organism: Streptomyces sp. ks84
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: MG |
 |
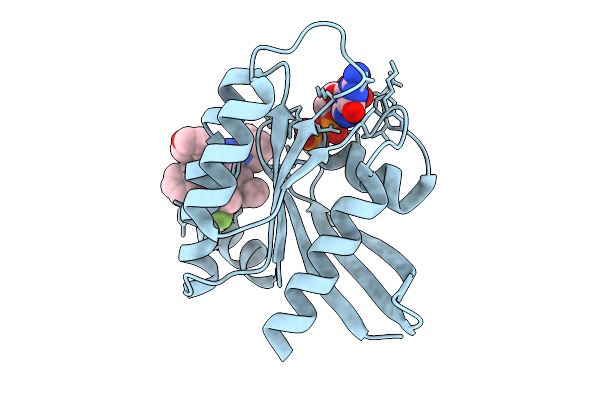
Organism: Streptomyces sp. ks84
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: SO4, EDO, FPS, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE Ligands: GDP, MG, A1L65 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE Ligands: A1L66, GDP, MG |
 |
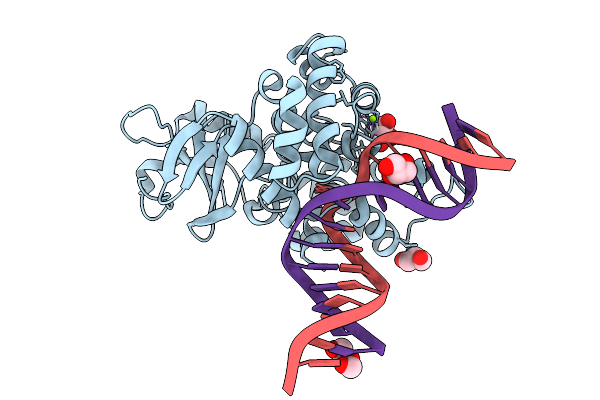
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: NUCLEAR PROTEIN Ligands: A1MAM, EDO, ACT |
 |
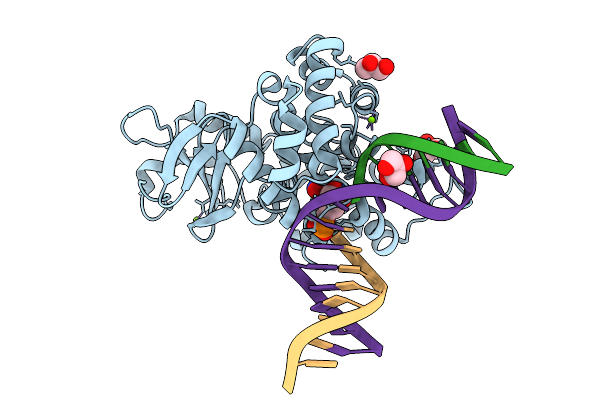
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Substrate 8-Oxoguanine Dna At Ph 8.0 Under 277 K
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: PEG, GOL, MG, NA |
 |
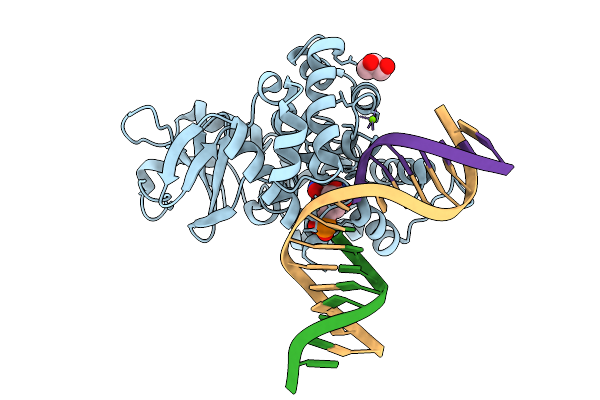
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 24 Hourss
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 2.5 Hours
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 298 K For 3 Weeks
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, GOL, MG, NA |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: PLANT PROTEIN |