Search Count: 99
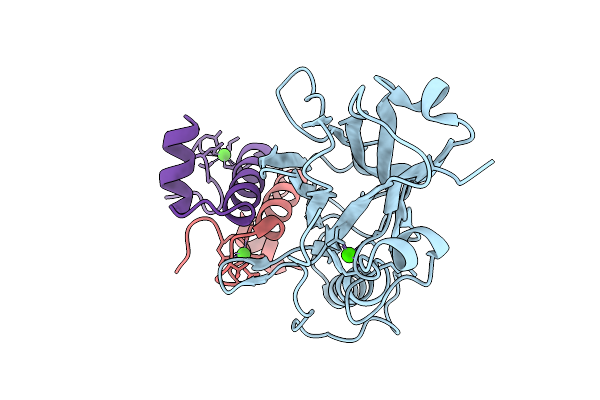 |
Ruminococcus Flavefaciens Coh-Doc Complex Between A Group 2 Dockerin And The Cohesin From Cell Surface Attached Scaffoldin Scae
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: PROTEIN BINDING Ligands: CA |
 |
Experimental Localization Of Metal-Binding Sites Reveals The Role Of Metal Ions In The Delafloxacin-Stabilized Streptococcus Pneumoniae Topoisomerase Iv Dna Cleavage Complex
Organism: Streptococcus pneumoniae, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-10-16 Classification: DNA BINDING PROTEIN Ligands: MPD, MG, K, CL, ACT, TE9 |
 |
Nucleant-Assisted 2.0 A Resolution Structure Of The Streptococcus Pneumoniae Topoisomerase Iv-V18Mer Dna Complex With The Novel Fluoroquinolone Delafloxacin
Organism: Streptococcus pneumoniae, Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-10-02 Classification: ISOMERASE Ligands: MG, CL, MPD, MLA, ACT, FMT, TE9 |
 |
High Resolution Structure Of The Streptococcus Pneumoniae Topoisomerase Iv-Complex With The V-Site 18Mer Dsdna And Novel Fluoroquinolone Delafloxacin
Organism: Streptococcus pneumoniae, Dna molecule
Method: X-RAY DIFFRACTION Release Date: 2024-10-02 Classification: ISOMERASE Ligands: MLA, ACT, MG, CL, MPD, TE9 |
 |
High Resolution Structure Of The Streptococcus Pneumoniae Topoisomerase Iv-Dna Complex With The Novel Fluoroquinolone Delafloxacin
Organism: Streptococcus pneumoniae, Dna molecule
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: ISOMERASE Ligands: MPD, MG, CL, TE9 |
 |
Proline Racemase (Pror) From The Gram-Positive Bacterium Acetoanaerobium Sticklandii From Isotropic Tetragonal Data At 3.15 A
Organism: Acetoanaerobium sticklandii dsm 519
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2023-07-05 Classification: ISOMERASE Ligands: PYC |
 |
Proline Racemase (Pror) From The Gram-Positive Bacterium Acetoanaerobium Sticklandii From Isotropic Orthorhombic Data At 3.59 A
Organism: Acetoanaerobium sticklandii dsm 519
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2023-07-05 Classification: ISOMERASE Ligands: PYC |
 |
Crystal Structure Of The Dimeric C-Terminal Big_2-Cbm56 Domains From Paenibacillus Illinoisensis (Bacillus Circulans Iam1165) Beta-1,3-Glucanase H
Organism: Paenibacillus illinoisensis
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: PG4, GOL, PG6, CL |
 |
Crystal Structure Of The Tetrameric C-Terminal Big_2-Cbm56 Domains From Paenibacillus Illinoisensis (Bacillus Circulans Iam1165) Beta-1,3-Glucanase H
Organism: Paenibacillus illinoisensis
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: PG4, PEG, GOL, CL, MG |
 |
Crystal Structure Of The Semet Octameric C-Terminal Big_2-Cbm56 Domains From Paenibacillus Illinoisensis (Bacillus Circulans Iam1165) Beta-1,3-Glucanase H
Organism: Paenibacillus illinoisensis
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-02-01 Classification: HYDROLASE Ligands: GOL, CL |
 |
Ruminococcus Flavefaciens Cohesin-Dockerin Structure: Dockerin From Scah Adaptor Scaffoldin In Complex With The Cohesin From Scae Anchoring Scaffoldin
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-11-02 Classification: PROTEIN BINDING Ligands: CA, GOL, SO4 |
 |
Structural And Functional Analysis Of The Proline Racemase (Pror) From The Gram-Positive Bacterium Acetoanaerobium Sticklandii
Organism: Acetoanaerobium sticklandii (strain atcc 12662 / dsm 519 / jcm 1433 / ncimb 10654)
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2022-08-10 Classification: ISOMERASE Ligands: PYC |
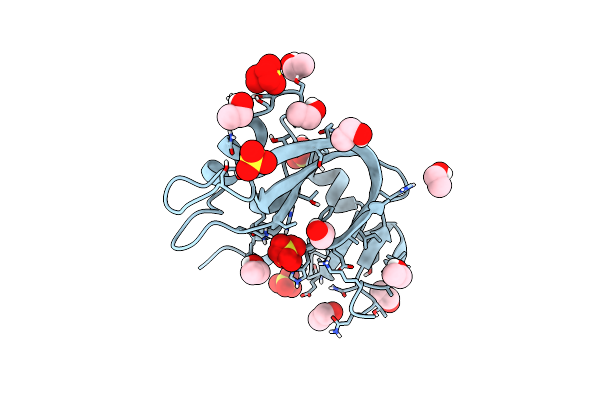 |
Ultra High Resolution X-Ray Structure Of Orthorhombic Bovine Pancreatic Ribonuclease At 100K
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2022-07-27 Classification: HYDROLASE Ligands: EOH, SO4 |
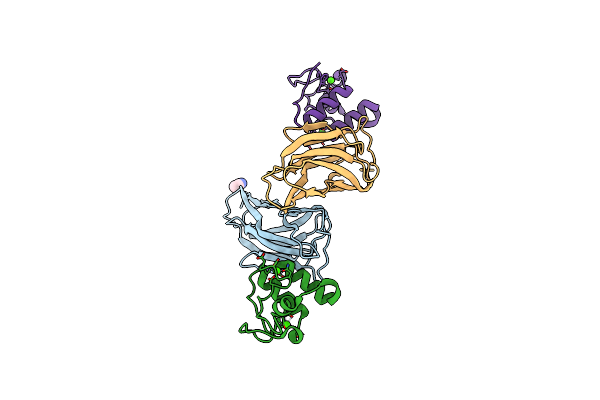 |
Crystal Structure Of Ruminococcus Flavefaciens' Type Iii Complex Containing The Fifth Cohesin From Scaffoldin B And The Dockerin From Scaffoldin A
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-02-28 Classification: PROTEIN BINDING Ligands: CCN, CA |
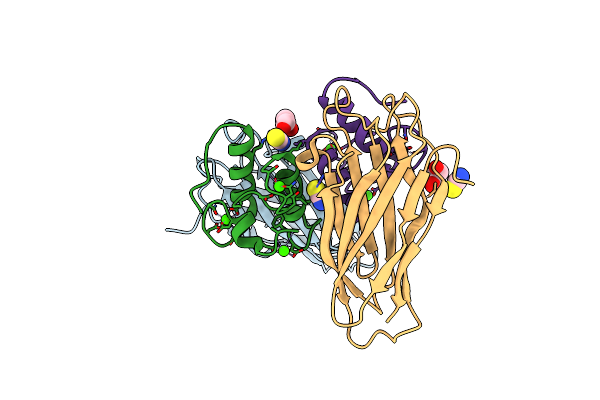 |
Crystal Structure Of The Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B In Complex With Cel5 Dockerin S15I, I16N Mutant
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-01-31 Classification: PROTEIN BINDING Ligands: CA, SCN, GOL |
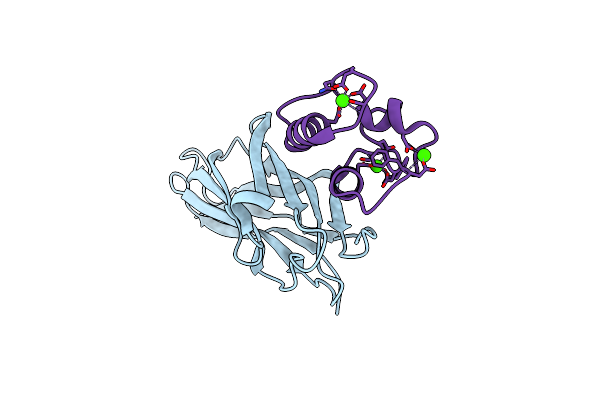 |
Crystal Structure Of The Sixth Cohesin From Acetivibrio Cellulolyticus' Scaffoldin B In Complex With Cel5 Dockerin S51I, L52N Mutant
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-01-31 Classification: CELL ADHESION Ligands: CA |
 |
Organism: Cupriavidus taiwanensis (strain dsm 17343 / bcrc 17206 / cip 107171 / lmg 19424 / r1)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-09-13 Classification: TOXIN |
 |
Crystal Structure Of The Cohscaa-Xdoccipb Type Ii Complex From Clostridium Thermocellum At 1.5Angstrom Resolution
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-09-06 Classification: CELL ADHESION Ligands: CA, SO4, GOL, EDO, PGE, P6G |
 |
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2017-07-05 Classification: PROTEIN BINDING Ligands: CA |
 |
Organism: Ruminococcus flavefaciens, Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-07-05 Classification: PROTEIN BINDING Ligands: CA, GOL |

