Search Count: 30
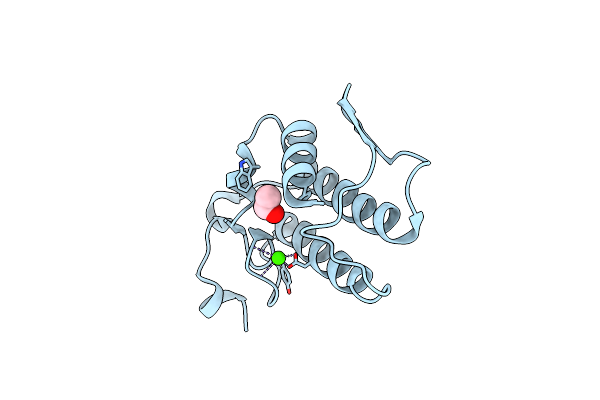 |
Crystal Structure Of Group I Phospholipase A2 At 2.3 A Resolution In 40% Ethanol Revealed The Critical Elements Of Hydrophobicity Of The Substrate-Binding Site
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-01-26 Classification: HYDROLASE Ligands: CA, EOH |
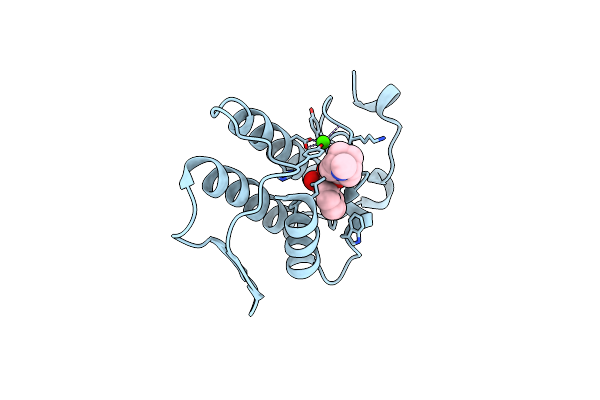 |
Crystal Structure Of The Complex Of Group 1 Phospholipase A2 With Atropin At 1.5 A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-11-17 Classification: HYDROLASE Ligands: CA, OIN |
 |
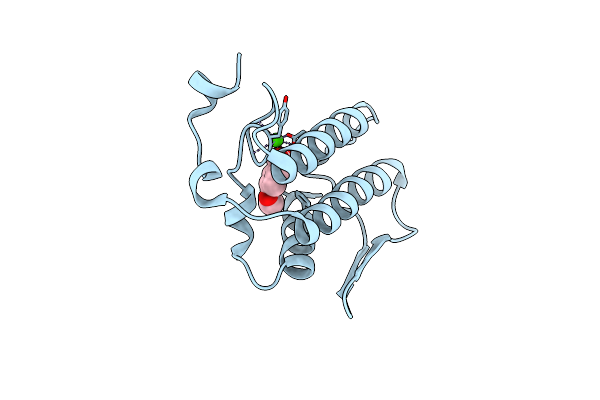
Crystal Structure Of The Complex Formed Between Phospholipase A2 With Beta-Amyloid Fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met At 1.8 A Resolution
Organism: Homo sapiens, Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-07-21 Classification: HYDROLASE Ligands: CA |
 |
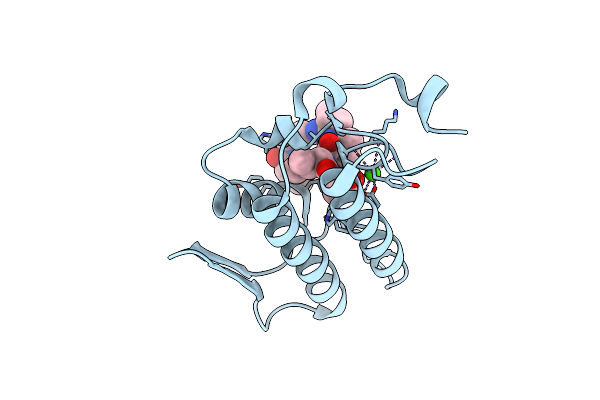
Crystal Structure Of The Complex Of Group I Phospholipase A2 With 4-Methoxy-Benzoicacid At 1.4A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-07-14 Classification: HYDROLASE Ligands: ANN, CA |
 |
Phospholipase A2 Prevents The Aggregation Of Amyloid Beta Peptides: Crystal Structure Of The Complex Of Phospholipase A2 With Octapeptide Fragment Of Amyloid Beta Peptide, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser At 2 A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2009-09-29 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of The Complex Formed Between Phospholipase A2 And A Hexapeptide Fragment Of Amyloid Beta Peptide, Lys-Leu-Val-Phe-Phe-Ala At 1.2 A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2009-09-29 Classification: HYDROLASE Ligands: CA |
 |
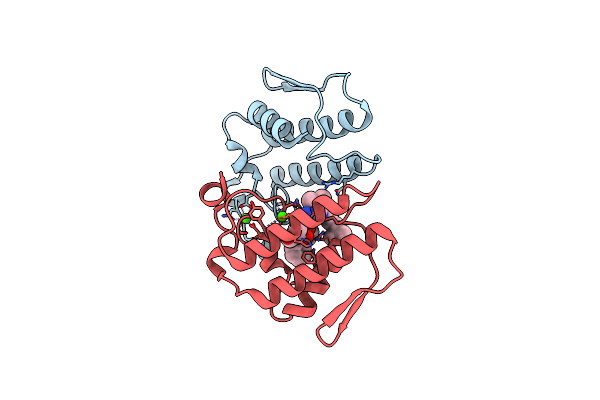
Crystal Structure Of The Complex Formed Between A New Isoform Of Phospholipase A2 With C-Terminal Amyloid Beta Heptapeptide At 2 A Resolution
Organism: Homo sapiens, Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2009-03-10 Classification: HYDROLASE Ligands: CA |
 |
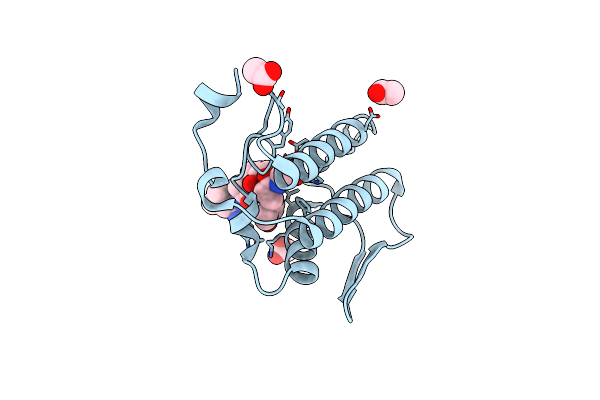
Design Of Specific Inhibitors Of Phospholipase A2: Crystal Structure Of The Complex Of Phospholipase A2 With Pentapeptide Leu-Val-Phe-Phe-Ala At 2.9 A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2007-10-09 Classification: HYDROLASE Ligands: CA |
 |
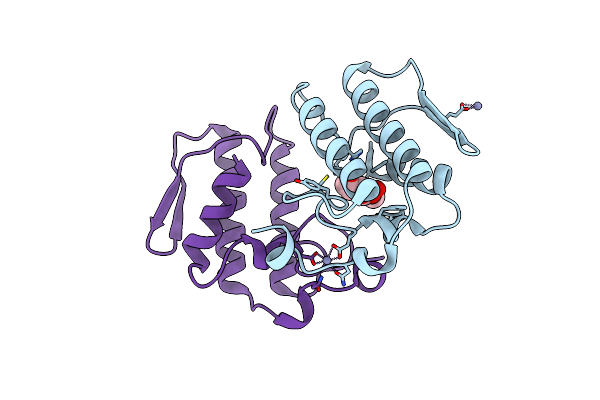
Crystal Structure Of The Complex Formed Beween A Group I Phospholipase A2 And Designed Penta Peptide Leu-Ala-Ile-Tyr-Ser At 2.6A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-06-21 Classification: HYDROLASE Ligands: ACT |
 |
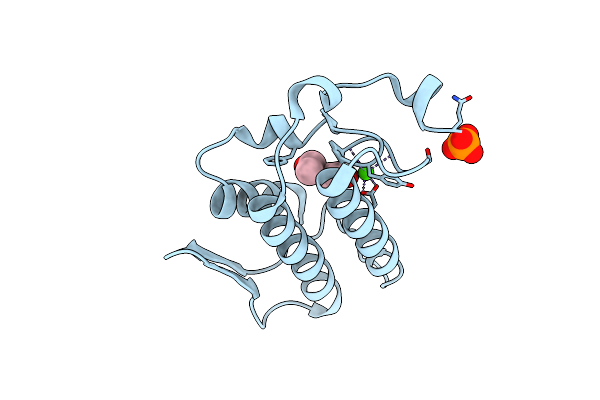
A New Form Of Catalytically Inactive Phospholipase A2 With An Unusual Disulphide Bridge Cys 32- Cys 49 Reveals Recognition For N-Acetylglucosmine
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-05-03 Classification: HYDROLASE Ligands: NAG, ZN |
 |
Crystal Structure Of A Novel Phospholipase A2 From Naja Naja Sagittifera With A Strong Anticoagulant Activity
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2005-05-03 Classification: HYDROLASE Ligands: CA, PO4, EOH |
 |
Structure Of Zinc Induced Heterodimer Of Two Calcium Free Isoforms Of Phospholipase A2 From Naja Naja Sagittifera At 2.7A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2005-03-15 Classification: HYDROLASE Ligands: ZN, ACY |
 |
Crystal Structure Of A Novel Phospholipase A2 From Naja Naja Sagittifera At 1.5 A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2005-03-08 Classification: HYDROLASE Ligands: CA, PO4, ACY |
 |
Interactions Of A Specific Non-Steroidal Anti-Inflammatory Drug (Nsaid) With Group I Phospholipase A2 (Pla2): Crystal Structure Of The Complex Formed Between Pla2 And Niflumic Acid At 2.5 A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-06-08 Classification: HYDROLASE Ligands: CA, NFL |
 |
Design Of Specific Inhibitors Of Phospholipase A2: Crystal Structure Of The Complex Formed Between Group I Phospholipase A2 And A Designed Pentapeptide Leu-Ala-Ile-Tyr-Ser At 2.6A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2004-05-04 Classification: HYDROLASE Ligands: ACT |
 |
Aspirin Induces Its Anti-Inflammatory Effects Through Its Specific Binding To Phospholipase A2: Crystal Structure Of The Complex Formed Between Phospholipase A2 And Aspirin At 1.9A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2004-04-27 Classification: HYDROLASE Ligands: AIN, CA |
 |
Crystal Structure Of An Acidic Phospholipase A2 From Naja Naja Sagittifera At 1.5 A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2004-04-20 Classification: HYDROLASE Ligands: CA, PO4, ACY |
 |
X-Ray Crystal Structure Of A Complex Formed Between Two Homologous Isoforms Of Phospholipase A2 From Naja Naja Sagittifera: Principle Of Molecular Association And Inactivation
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-02-10 Classification: HYDROLASE Ligands: CA, PO4, ACY |
 |
Structure-Based Design Of Potent And Selective Inhibitors Of Phospholipase A2: Crystal Structure Of The Complex Formed Between Phosholipase A2 From Naja Naja Sagittifera And A Designed Peptide Inhibitor At 1.9 A Resolution
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-09-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA |
 |
Crystal Structure Of A Phopholipase A2 Monomer With Isoleucine At Second Position
Organism: Naja sagittifera
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2003-06-10 Classification: HYDROLASE |

