Search Count: 28
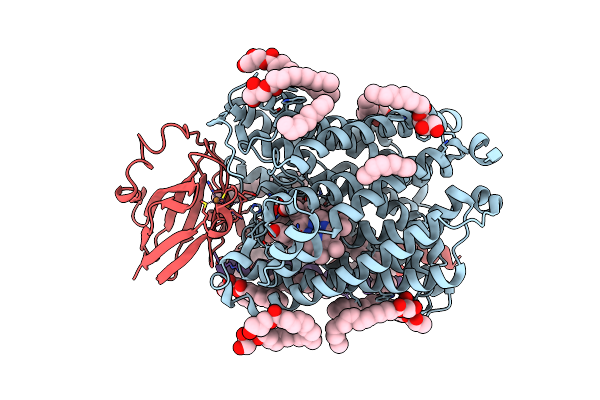 |
Serial Femtosecond Crystallography Structure Of Co Bound Ba3- Type Cytochrome C Oxidase Without Pump Laser Irradiation
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-15 Classification: ELECTRON TRANSPORT Ligands: CU, HEM, HAS, OLC, CMO, CUA |
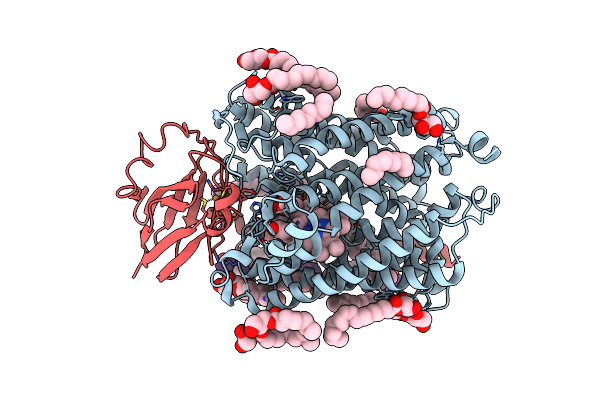 |
Serial Femtosecond Crystallography Structure Of Photo Dissociated Co From Ba3- Type Cytochrome C Oxidase Determined By Extrapolation Method
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-15 Classification: ELECTRON TRANSPORT Ligands: CU, HEM, HAS, OLC, CMO, CUA |
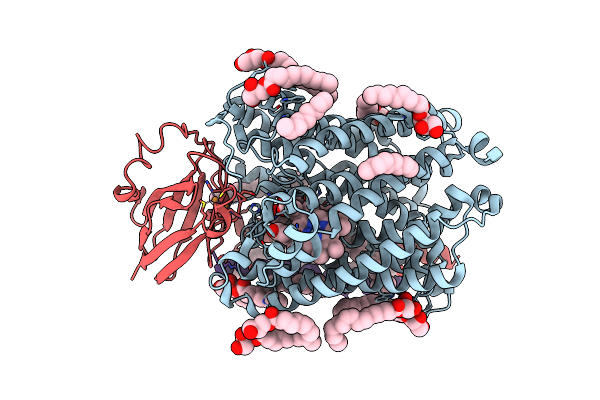 |
Serial Femtosecond Crystallography Structure Of Co Bound Ba3- Type Cytochrome C Oxidase At 2 Milliseconds After Irradiation By A 532 Nm Laser
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-08-16 Classification: ELECTRON TRANSPORT Ligands: CU, HEM, HAS, OLC, CMO, CUA |
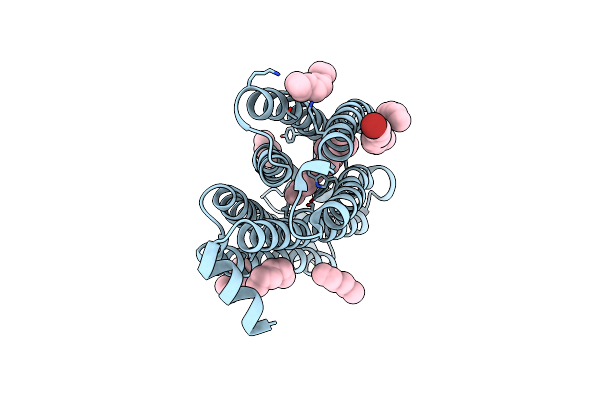 |
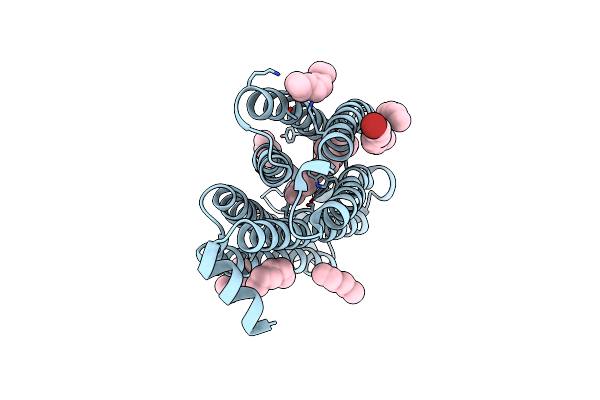
Time-Resolved Serial Femtosecond Crystallography Structure Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3: Resting State Structure With Bromide Ion
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, D10, BR |
 |
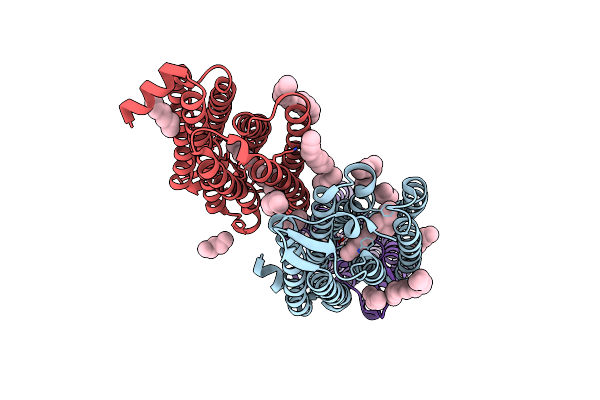
Time-Resolved Serial Femtosecond Crystallography Structure Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3 : Structure Obtained 1 Msec After Photoexcitation With Bromide Ion
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, D10, BR |
 |
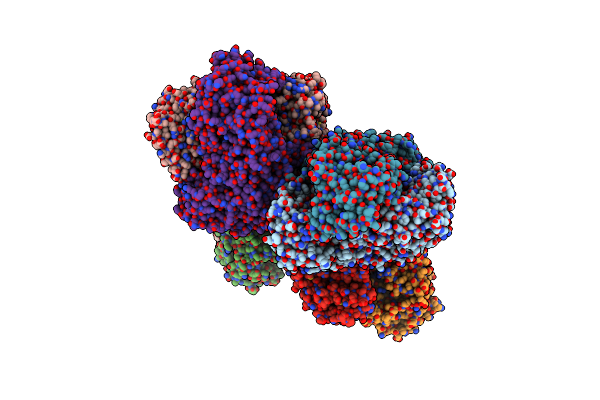
Anion Free Form Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3, Structure Determined By Serial Femtosecond Crystallography At Sacla
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, DD9, C14, R16, OCT, CL |
 |
Ambient Temperature Structure Of Bifidobacterium Longum Phosphoketolase With Thiamine Diphosphate
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-02 Classification: LYASE Ligands: TPP, CA, LMR, MLA, SIN |
 |
Ambient Temperature Structure Of Bifidobacgterium Longum Phosphoketolase With Thiamine Diphosphate And Phosphoenol Pyuruvate
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-06-02 Classification: LYASE Ligands: PEP, TPP, CA |
 |
 |
 |
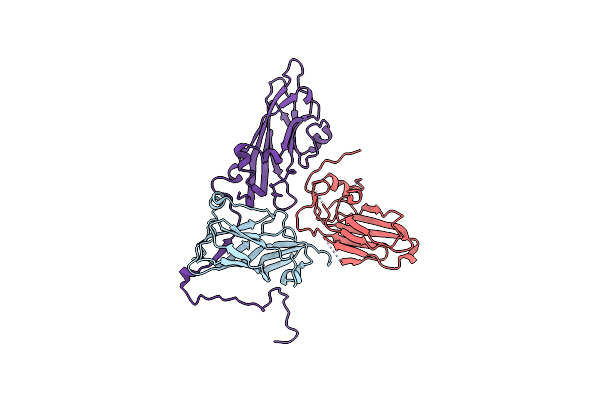 |
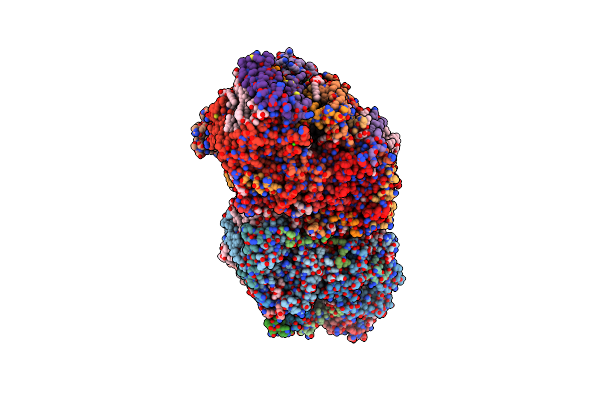
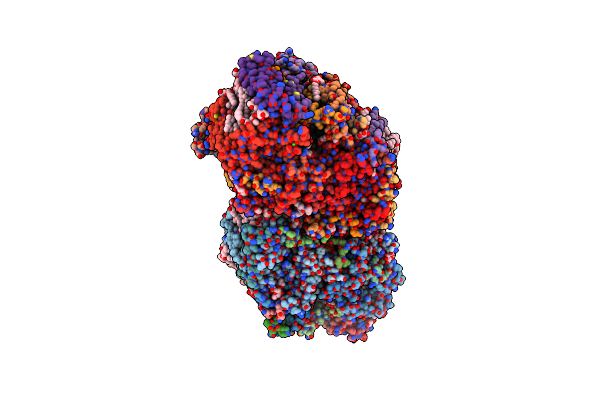
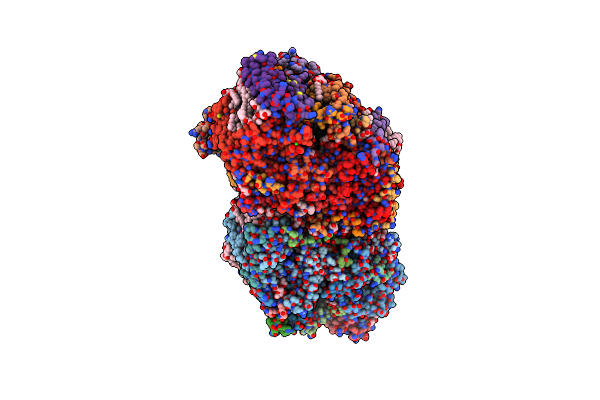
Organism: Apple latent spherical virus
Method: ELECTRON MICROSCOPY Release Date: 2020-11-04 Classification: STRUCTURAL PROTEIN |
 |
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-03-06 Classification: TRANSFERASE |
 |
Structure Of Biotin Carboxyl Carrier Protein From Pyrococcus Horikoshi Ot3 (Delta N79) Wild Type
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-08-30 Classification: TRANSFERASE |
 |
Structure Of Biotin Carboxyl Carrier Protein From Pyrococcus Horikoshi Ot3 (Delta N79) A138I Mutant
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-08-30 Classification: TRANSFERASE |
 |
Structure Of Biotin Carboxyl Carrier Protein From Pyrococcus Horikoshi Ot3 (Delta N79) A138Y Mutant
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-08-30 Classification: TRANSFERASE |
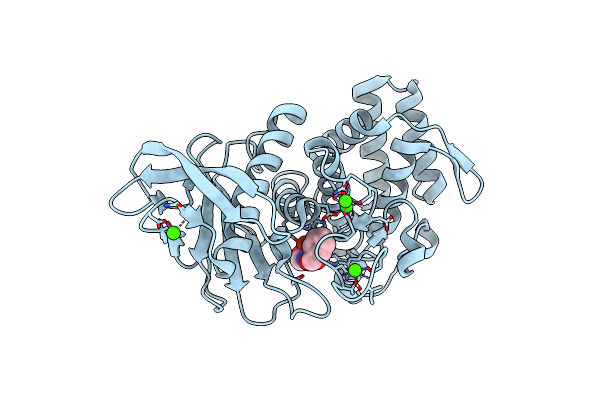 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: NX6, ZN, CA |
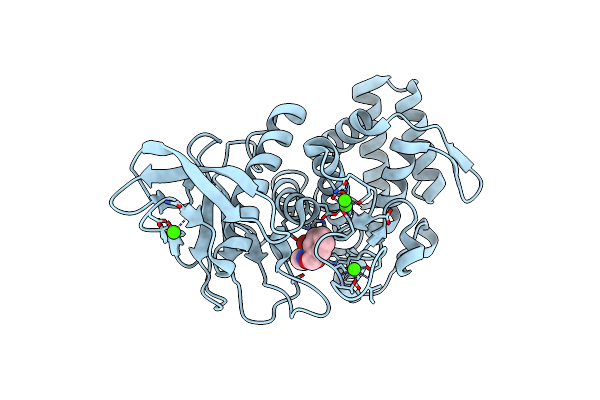 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: NX6, ZN, CA |
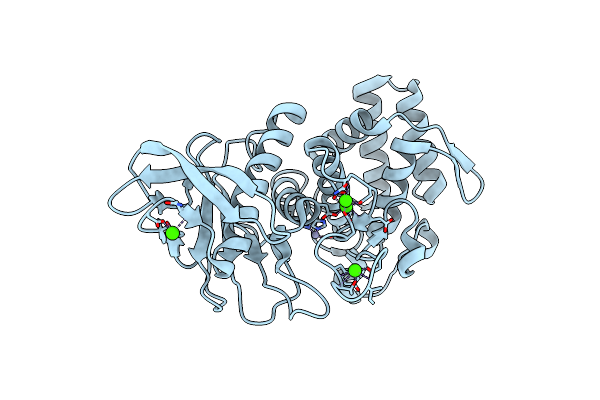 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: ZN, CA |
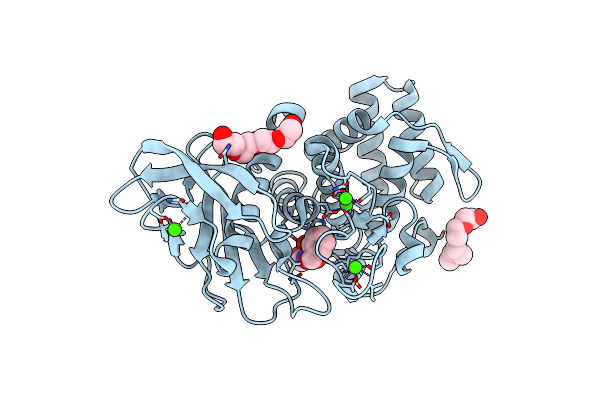 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: NX6, ZN, CA, PG4 |

