Search Count: 37
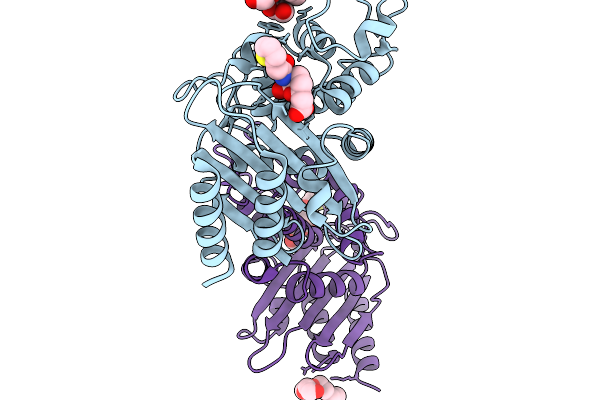 |
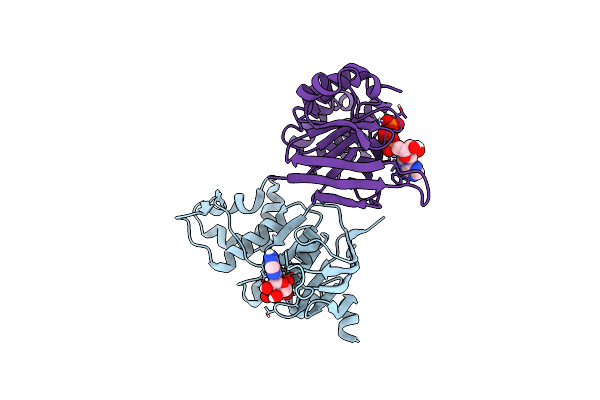
Crystal Structure Of Vaborbactam In Complex With Sme-1 Class A Carbapenemase
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE Ligands: 4D6, PG4, EDO |
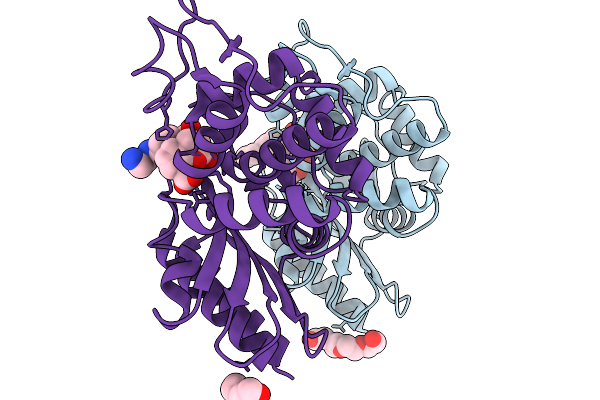 |
Crystal Structure Of Taniborbactam In Complex With Sme-1 Class A Carbapenemase
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE Ligands: KJK, PG4 |
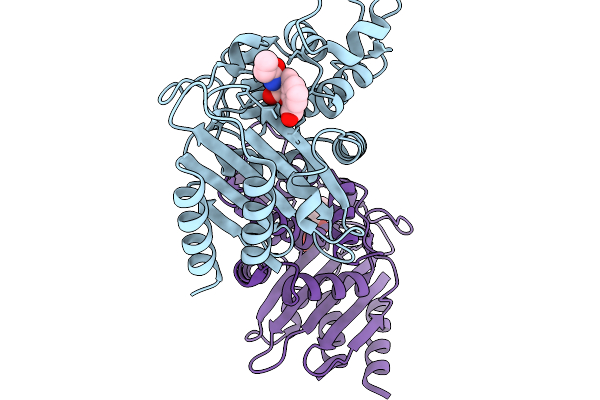 |
Crystal Structure Of Ledaborbactam In Complex With Sme-1 Class A Carbapenemase
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE Ligands: A1MBD, CL |
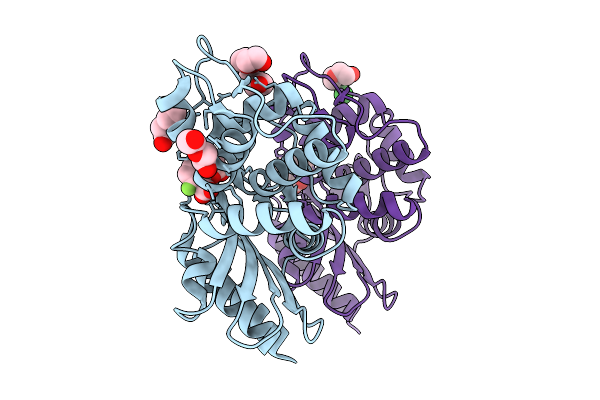 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: A1MAF, PEG, PG4, CL, NA |
 |
Crystal Structure Of Xeruborbactam (Qpx7728) In Complex With Sme-1 Class A Carbapenemase
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: EDO, RM9, CL |
 |
Single Particle Cryoem Structure Of The Pf80S Ribosome In Non-Rotated Pre State (Nrt A-P-E)
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: RIBOSOME |
 |
Single Particle Cryoem Structure Of The Pf80S Ribosome In The Post State (Nrt With P- And E-Site Trna)
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: RIBOSOME |
 |
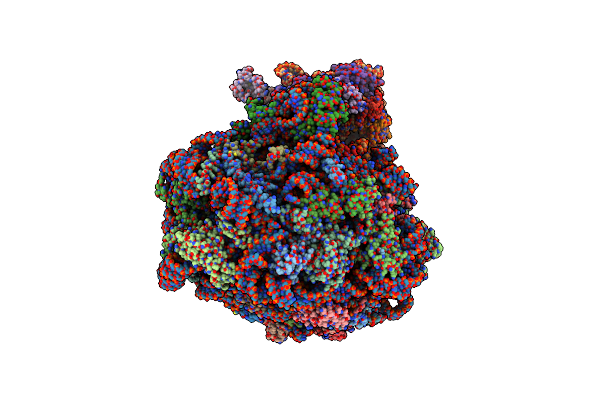
Single Particle Cryoem Structure Of The Pf80S Ribosome In The Unloaded State (Nrt With E-Site Trna)
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: RIBOSOME |
 |
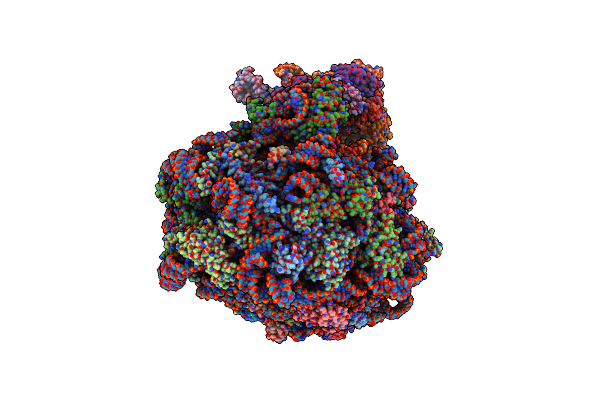
Single Particle Cryoem Structure Of The Pf80S Ribosome In The Rotated-2 Pre State (Rt State With P And E-Site Trna)
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: RIBOSOME |
 |
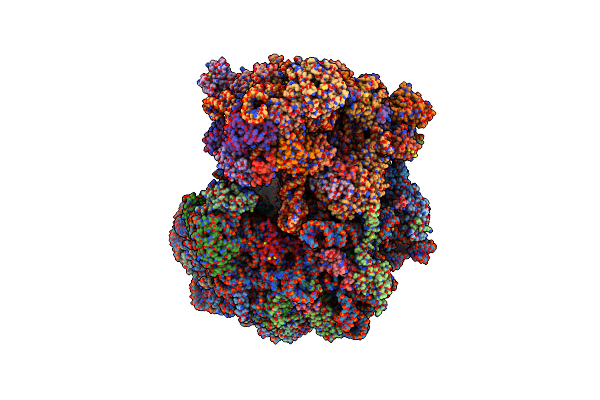
Single Particle Cryoem Structure Of The Pf80S Ribosome In Rotated State With E-Site Trna
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: RIBOSOME |
 |
Subtomogram Averaged Consensus Structure Of The Malarial 80S Ribosome In Plasmodium Falciparum-Infected Human Erythrocytes
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: RIBOSOME |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2021-04-07 Classification: HYDROLASE Ligands: ATP, MG |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2016-06-29 Classification: TRANSFERASE Ligands: PEG |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-06-29 Classification: TRANSFERASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2016-06-29 Classification: TRANSFERASE |
 |
Structure And Mechanism Of A Dehydratase/Decarboxylase Enzyme Couple Involved In Polyketide Beta-Branching
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-05-06 Classification: LYASE Ligands: EDO, GOL |
 |
Structure And Mechanism Of A Dehydratase/Decarboxylase Enzyme Couple Involved In Polyketide Beta-Branching
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2015-05-06 Classification: LYASE Ligands: GOL, NA, EPE |
 |
Structure And Mechanism Of A Dehydratase/Decarboxylase Enzyme Couple Involved In Polyketide Beta-Branching
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-05-06 Classification: LYASE Ligands: GOL |
 |
Structure And Mechanism Of A Dehydratase/Decarboxylase Enzyme Couple Involved In Polyketide Beta-Branching
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2015-05-06 Classification: LYASE Ligands: EDO, NA |
 |
Structure And Mechanism Of A Dehydratase/Decarboxylase Enzyme Couple Involved In Polyketide Beta-Branching
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-05-06 Classification: LYASE Ligands: GOL, PO4 |

