Search Count: 33
 |
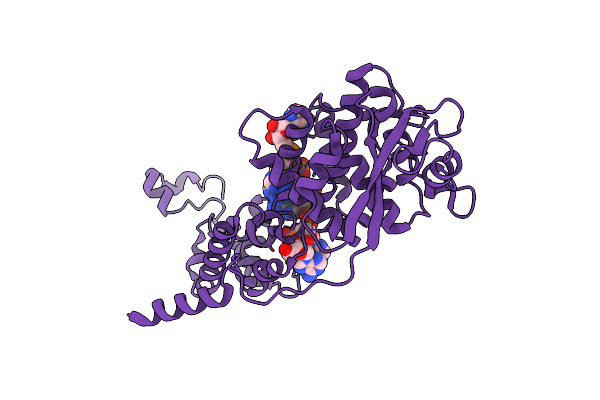
Cryo-Em Structure Of The Spike Glycoprotein From Bat Sars-Like Coronavirus (Bat Sl-Cov) Wiv1 In Locked State
Organism: Bat sars-like coronavirus wiv1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: VIRAL PROTEIN Ligands: EIC, NAG |
 |
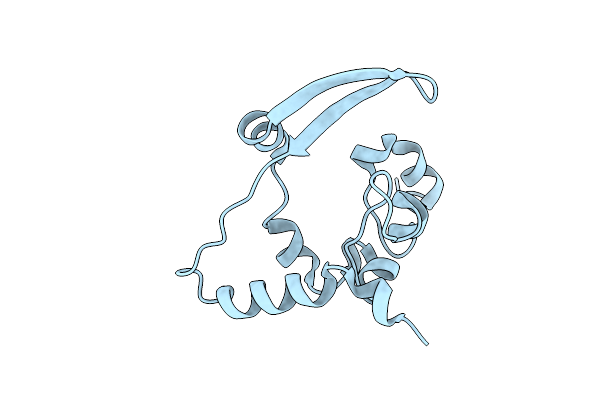
Organism: Morbillivirus sp.
Method: ELECTRON MICROSCOPY Release Date: 2021-06-23 Classification: VIRAL PROTEIN |
 |
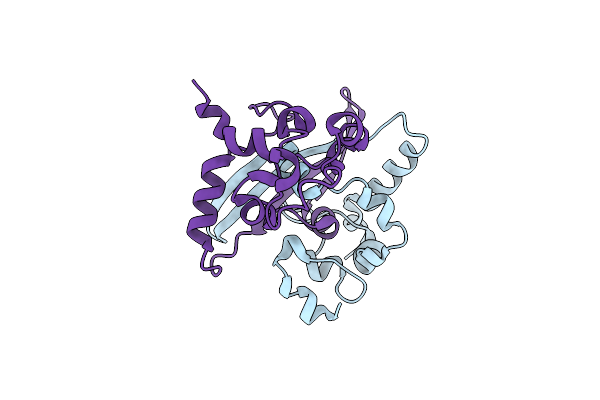
Crystal Structure Of C-Terminal Dimerization Domain Of Nucleocapsid Phosphoprotein From Sars-Cov-2, Crystal Form Ii
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2020-07-01 Classification: VIRAL PROTEIN |
 |
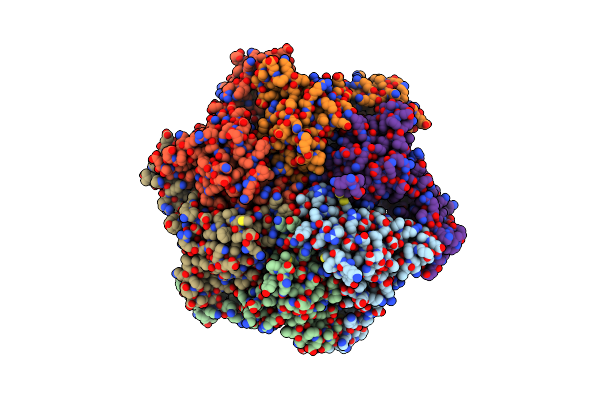
1.45 Angstrom Resolution Crystal Structure Of C-Terminal Dimerization Domain Of Nucleocapsid Phosphoprotein From Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2020-05-20 Classification: VIRAL PROTEIN |
 |
Organism: Archaeoglobus fulgidus (strain atcc 49558 / vc-16 / dsm 4304 / jcm 9628 / nbrc 100126)
Method: ELECTRON MICROSCOPY Release Date: 2018-12-26 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Archaeoglobus fulgidus (strain atcc 49558 / vc-16 / dsm 4304 / jcm 9628 / nbrc 100126)
Method: ELECTRON MICROSCOPY Release Date: 2018-12-26 Classification: HYDROLASE |
 |
Organism: Archaeoglobus fulgidus dsm 4304
Method: ELECTRON MICROSCOPY Release Date: 2018-12-26 Classification: HYDROLASE |
 |
Organism: Archaeoglobus fulgidus (strain atcc 49558 / vc-16 / dsm 4304 / jcm 9628 / nbrc 100126)
Method: ELECTRON MICROSCOPY Release Date: 2018-12-26 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Archaeoglobus fulgidus (strain atcc 49558 / vc-16 / dsm 4304 / jcm 9628 / nbrc 100126)
Method: ELECTRON MICROSCOPY Release Date: 2018-12-26 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Archaeoglobus fulgidus dsm 4304
Method: ELECTRON MICROSCOPY Release Date: 2018-12-26 Classification: HYDROLASE Ligands: MG, ATP, ADP |
 |
Organism: Archaeoglobus fulgidus (strain atcc 49558 / vc-16 / dsm 4304 / jcm 9628 / nbrc 100126)
Method: ELECTRON MICROSCOPY Release Date: 2018-12-26 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Archaeoglobus fulgidus dsm 4304
Method: ELECTRON MICROSCOPY Release Date: 2018-12-26 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Ebola virus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-10-10 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of The Oligomerization Domain Of Vp35 From Ebola Virus, Mercury Derivative
Organism: Zaire ebolavirus
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2018-10-10 Classification: VIRAL PROTEIN Ligands: HG |
 |
Organism: Reston ebolavirus
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2018-10-10 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of The Oligomerization Domain Of Vp35 From Reston Virus, Mercury Derivative
Organism: Reston ebolavirus
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2018-10-10 Classification: VIRAL PROTEIN Ligands: MBO |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-08-29 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-08-22 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-08-22 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-08-22 Classification: HYDROLASE Ligands: ADP, MG, ATP |

