Search Count: 52
 |
Organism: Microchaete diplosiphon
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-05-15 Classification: SIGNALING PROTEIN Ligands: CYC |
 |
Neutron Structure Of Bacillus Thermoproteolyticus Ferredoxin At Room Temperature
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.45 Å, 1.60 Å Release Date: 2024-02-07 Classification: ELECTRON TRANSPORT Ligands: SF4 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2023-02-22 Classification: HYDROLASE |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: TLA, GOL |
 |
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2022-10-26 Classification: CONTRACTILE PROTEIN Ligands: ANP, MG, PO4, EDO, CA |
 |
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2022-10-26 Classification: CONTRACTILE PROTEIN Ligands: ADP, PO4, MG, EDO, CA |
 |
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-10-26 Classification: CONTRACTILE PROTEIN Ligands: ADP, MG, EDO, CA |
 |
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-10-26 Classification: CONTRACTILE PROTEIN Ligands: ATP, CA, ACT, NA, EDO |
 |
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-10-26 Classification: CONTRACTILE PROTEIN Ligands: ADP, CA, ACT, NA, EDO, PO4 |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-09-07 Classification: PLANT PROTEIN Ligands: TLA, GOL |
 |
Organism: Microchaete diplosiphon
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-05-05 Classification: SIGNALING PROTEIN Ligands: CYC, MG |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2021-01-27 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-01-20 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: GA4, AKG, GOL, SO4 |
 |
Organism: Oryza sativa subsp. indica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-13 Classification: BIOSYNTHETIC PROTEIN Ligands: AKG, IAC, GOL |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: FOL, NAP |
 |
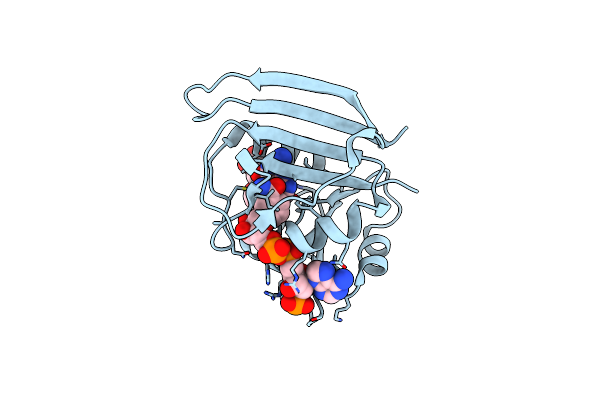
High-Pressure Crystal Structure Analysis Of M20 Loop Closed Dhfr At 220 Mpa
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: FOL, NAP |
 |
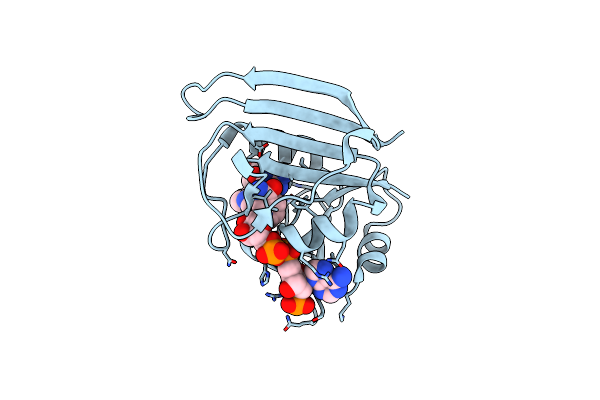
High-Pressure Crystal Structure Analysis Of M20 Loop Closed Dhfr At 400 Mpa
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: FOL, NAP |
 |
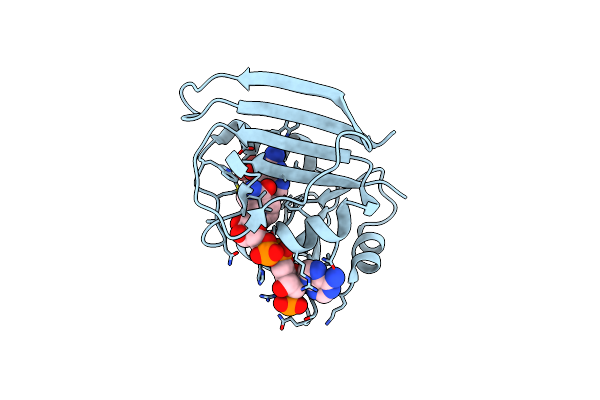
High-Pressure Crystal Structure Analysis Of M20 Loop Closed Dhfr At 650 Mpa
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: FOL, NAP |
 |
High-Pressure Crystal Structure Analysis Of M20 Loop Closed Dhfr At 800 Mpa
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: FOL, NAP |

