Search Count: 817
All
Selected
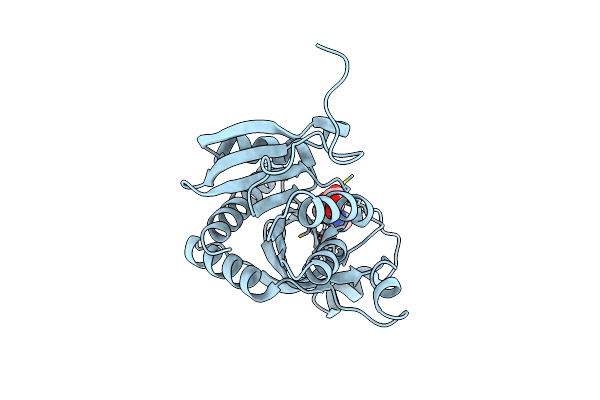 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Inosine Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-01-22 Classification: TRANSFERASE Ligands: NOS, CL |
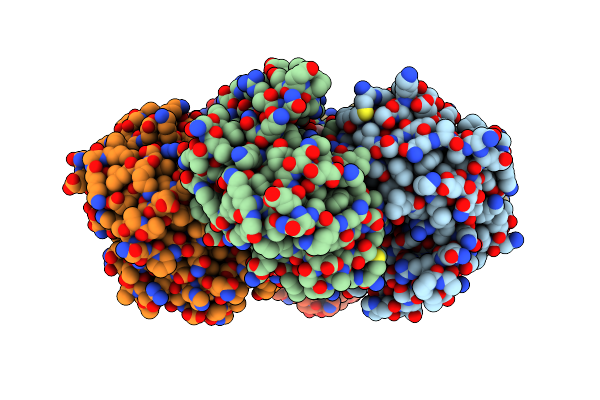 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (C2 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-25 Classification: TRANSFERASE Ligands: GOL |
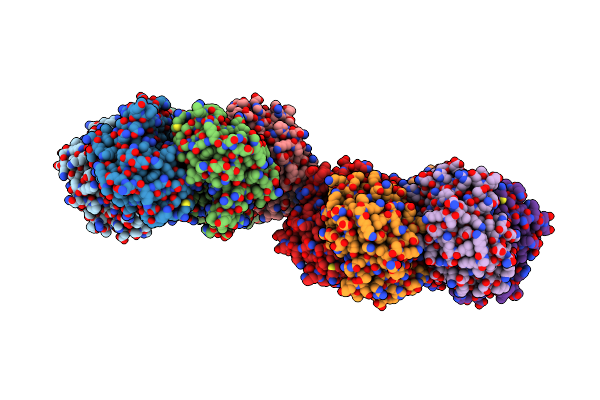 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (P21 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2024-12-25 Classification: TRANSFERASE Ligands: FLC |
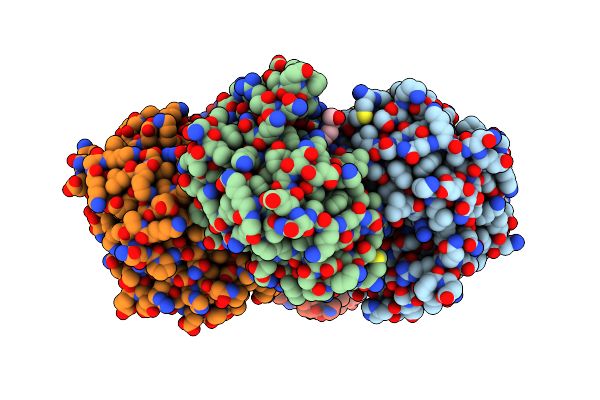 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Hepes Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-12-25 Classification: TRANSFERASE Ligands: GOL, EPE |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Adenosine And Phosphate Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: CL, ADN, PO4 |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Phosphate Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: GOL, PO4 |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Glycerol Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: GOL |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Mannose Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: GOL, MAN |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Citrate Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: CIT |
 |
Crystal Structure Of Apo Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (H32 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-12-18 Classification: TRANSFERASE |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Phosphate/Adenine Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: CL, PO4, ADE |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Adenosine And Glycine Complex)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: GLY, CL, ADN |
 |
Wild Type Purine Nucleoside Phosphorylase From E.Coli In Complex With N2,3-Etheno-2-Aminopurine
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-10-30 Classification: TRANSFERASE Ligands: GOL, A1IEC, PO4, SO4 |
 |
D204N Mutant Of Purine Nucleoside Phosphorylase From E.Coli In Complex With N2,3-Etheno-2-Aminopurine
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2024-10-30 Classification: TRANSFERASE Ligands: PO4, A1IEC |
 |
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-01-17 Classification: TRANSFERASE/INHIBITOR Ligands: XUH, EDO, NA, PO4 |
 |
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: HPA, ACT |
 |
Structure Of K. Lactis Pnp Bound To Transition State Analog Dadme-Immucillin H And Sulfate
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: DIH, SO4, GOL |
 |
Structure Of K. Lactis Pnp S42E Variant Bound To Transition State Analog Dadme-Immucillin G And Sulfate
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: IM5, SO4, GUN |
 |
Structure Of K. Lactis Pnp S42E-H98R Variant Bound To Transition State Analog Dadme-Immucillin G And Sulfate
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: IM5, SO4 |
 |
Structure Of Bacteroides Fragilis Pnp Bound To Transition State Analog Immucillin H And Sulfate
Organism: Bacteroides fragilis nctc 9343
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: IMH, SO4 |

