Search Count: 69
 |
Organism: Thalassospira sp.
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: EDO, NPO, SO4, 1PE |
 |
Crystal Structure Of A Sulfotransferase S4 From In Complex With Pap And Pnp
Organism: Bambusicola thoracicus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-01-15 Classification: TRANSFERASE Ligands: NPO, A3P |
 |
Organism: Bambusicola thoracicus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-12-25 Classification: TRANSFERASE Ligands: NPO, PPS, 4NS |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With P-Nitrophenol
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: EDO, NPO, ACE |
 |
Crystal Structure Of Dehaloperoxidase A In Complex With Substrate 4-Nitrophenol
Organism: Amphitrite ornata
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-07-17 Classification: OXIDOREDUCTASE Ligands: HEM, NPO, GOL, SO4 |
 |
Structure Of Kudzu 2-Hydroxyisoflavanone Dehydratase In Complex With P-Nitrophenol
Organism: Pueraria montana var. lobata
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2022-11-30 Classification: PLANT PROTEIN Ligands: NPO |
 |
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2022-03-09 Classification: HYDROLASE Ligands: NAG, BGC, GAL, NPO, CO |
 |
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-03-09 Classification: HYDROLASE Ligands: NAG, NPO, CO |
 |
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-02-23 Classification: HYDROLASE Ligands: GOL, EDO, 6NA, DMS, CCN, D8F, SO4, DIO, BUA, NPO, NA |
 |
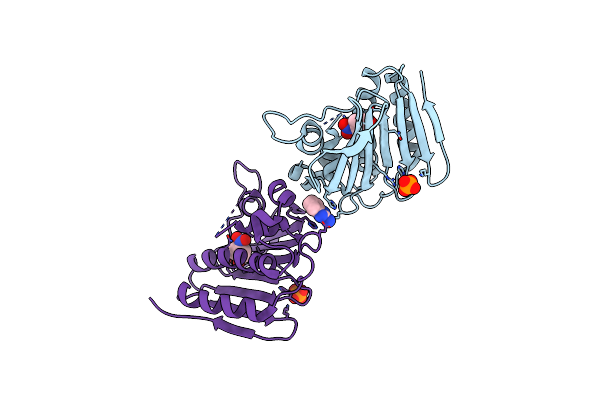
Enterococcus Faecalis Gh31 Alpha-N-Acetylgalactosaminidase D455A In Complex With P-Nitrophenyl Alpha-N-Acetylgalactosaminide
Organism: Enterococcus faecalis atcc 10100
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2021-12-08 Classification: HYDROLASE Ligands: A2G, NPO, EDO, 1PE |
 |
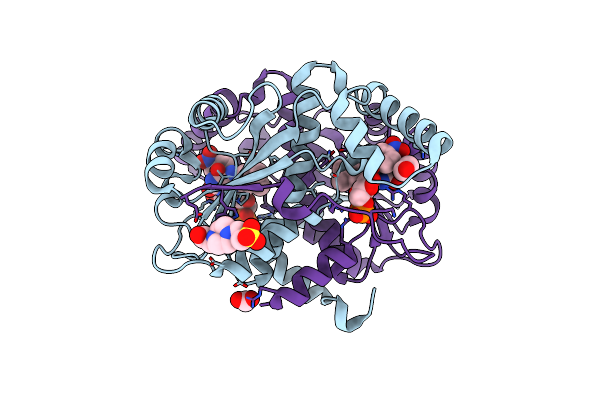
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-10-27 Classification: ISOMERASE Ligands: AX7, NPO, PO4 |
 |
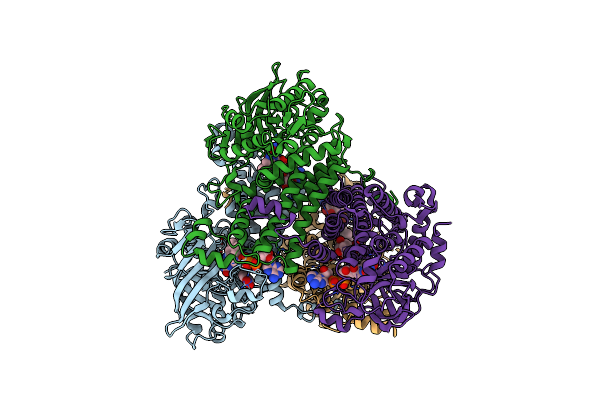
Crystal Structure Of Putative Nad(P)H-Flavin Oxidoreductase From Haemophilus Influenzae R2846 In Complex With 4-Nitrophenol
Organism: Haemophilus influenzae r2846
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: NPO, EDO, FMT, FMN, EPE |
 |
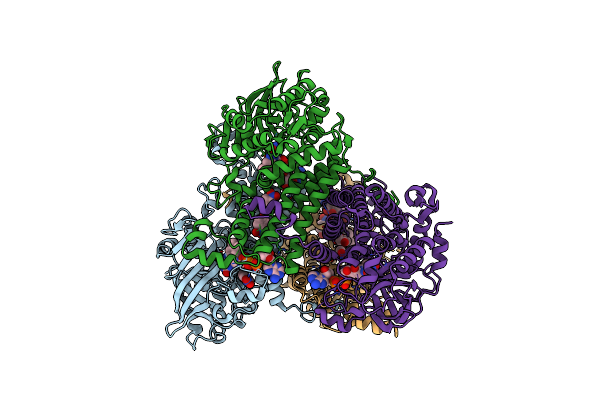
Crystal Structure Of A Flavin-Dependent Monooxygenase Hada Wild Type Complexed With Reduced Fad And 4-Nitrophenol
Organism: Ralstonia pickettii dtp0602
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: FDA, NPO |
 |
Crystal Structure Of A Flavin-Dependent Monooxygenase Hada F441V Mutant Complexed With Reduced Fad And 4-Nitrophenol
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: FDA, NPO |
 |
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-07-14 Classification: HYDROLASE Ligands: NPO, 6NA, PEG, EDO, DMS, DIO, GOL, SO4, PGE, NA, MES |
 |
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-07-14 Classification: HYDROLASE Ligands: D8F, DMS, GOL, EDO, DIO, NPO, SO4 |
 |
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-07-14 Classification: HYDROLASE Ligands: GOL, EDO, 6NA, DMS, CCN, D8F, SO4, DIO, BUA, NPO, NA |
 |
Crystal Structure Of 3-O-Sulfotransferase Isoform 3 In Complex With 8Mer Oligosaccharide With 6S Sulfation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-06-23 Classification: TRANSFERASE Ligands: NPO, A3P, NA, EDO |
 |
Chaetomium Thermophilum Fad-Dependent Oxidoreductase In Complex With 4-Nitrophenol
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-05-26 Classification: OXIDOREDUCTASE Ligands: FDA, NAG, NPO, FMT, ACT, PG4, PGE, MG, NA |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-03-03 Classification: HYDROLASE Ligands: FMT, NPO, ACT |

