Search Count: 13
 |
Pseudomonas Aeruginosa Dna Gyrase B 24Kda Atpase Subdomain Complexed With Novobiocin
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2022-10-05 Classification: DNA BINDING PROTEIN Ligands: NOV, MRD, MES |
 |
Acinetobacter Baumannii Dna Gyrase B 23Kda Atpase Subdomain Complexed With Novobiocin
Organism: Acinetobacter baumannii 1419130
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-09-28 Classification: DNA BINDING PROTEIN Ligands: NOV, EDO |
 |
Lc3A In Complex With (3R,4S,5R,6R)-5-Hydroxy-6-((4-Hydroxy-3-(4-Hydroxy-3-Isopentylbenzamido)-8-Methyl-2-Oxo-2H-Chromen-7-Yl)Oxy)-3-Methoxy-2,2-Dimethyltetrahydro-2H-Pyran-4-Yl Carbamate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: NOV, EDO |
 |
Organism: Mycolicibacterium thermoresistibile
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-08-12 Classification: DNA BINDING PROTEIN Ligands: NOV, ZN, EDO |
 |
Mycobacterium Smegmatis Gyrb 22Kda Atpase Sub-Domain In Complex With Novobiocin
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-08-12 Classification: DNA BINDING PROTEIN Ligands: NOV, EDO, ACT, NA |
 |
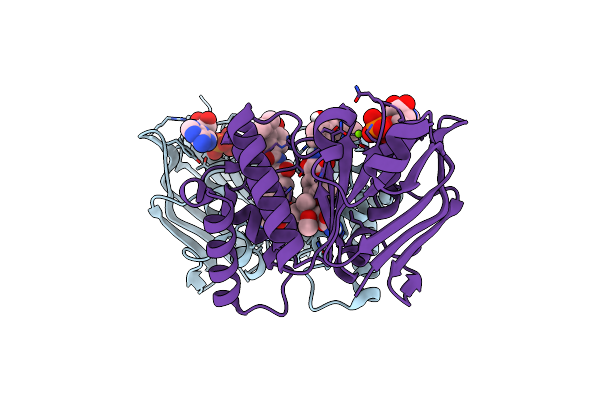
Organism: Enterobacter cloacae subsp. cloacae (strain atcc 13047 / dsm 30054 / nbrc 13535 / ncdc 279-56), Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-03-27 Classification: LIPID TRANSPORT Ligands: SO4, CL, NOV |
 |
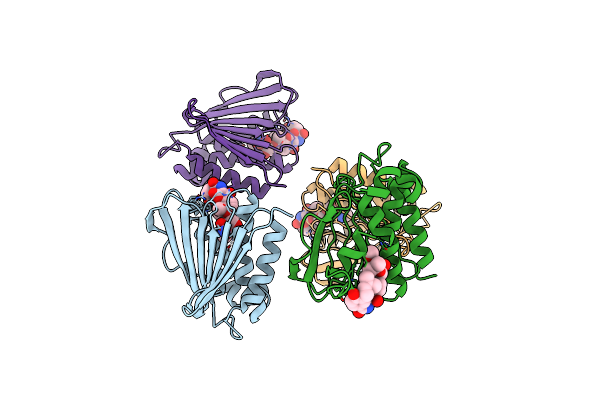
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-12-06 Classification: lipid transport/activator Ligands: ADP, MG, NOV |
 |
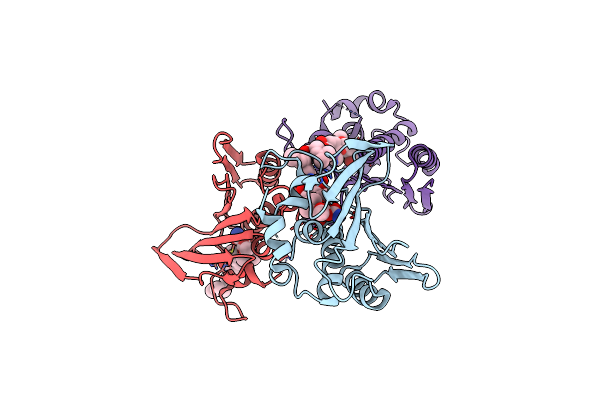
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-07-30 Classification: ISOMERASE Ligands: NOV |
 |
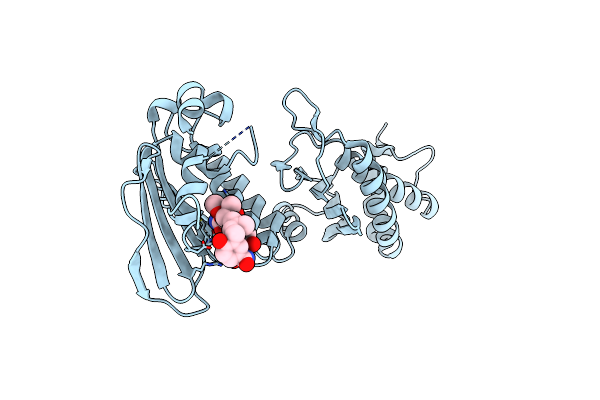
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-07-16 Classification: ISOMERASE Ligands: NOV |
 |
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2011-02-16 Classification: ISOMERASE/ANTIBIOTIC Ligands: NOV |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-05-04 Classification: ISOMERASE Ligands: NOV |
 |
Crystal Structure Of The 43K Atpase Domain Of Thermus Thermophilus Gyrase B In Complex With Novobiocin
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-06-03 Classification: ISOMERASE Ligands: NOV, FMT |
 |
Novobiocin-Resistant Mutant (R136H) Of The N-Terminal 24 Kda Fragment Of Dna Gyrase B Complexed With Novobiocin At 2.3 Angstroms Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1998-05-20 Classification: TOPOISOMERASE Ligands: NOV |

