Search Count: 29
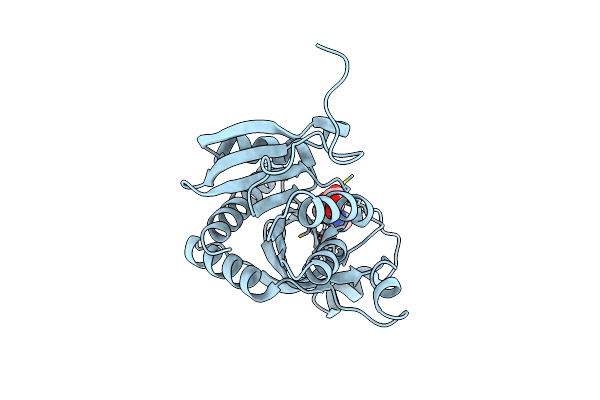 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Inosine Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-01-22 Classification: TRANSFERASE Ligands: NOS, CL |
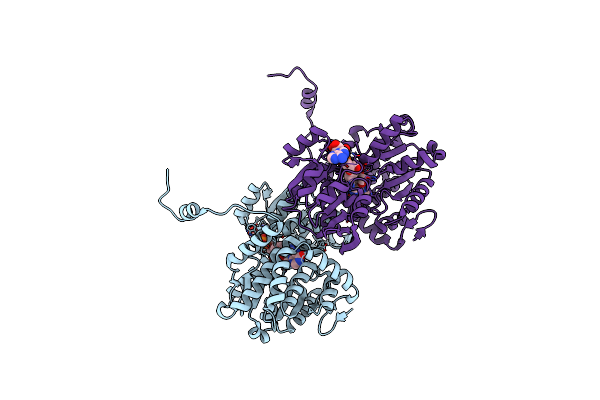 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase Treated At 368 K From Pyrococcus Furiosus In Complex With Inosine
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-08-21 Classification: HYDROLASE Ligands: NOS, NAD |
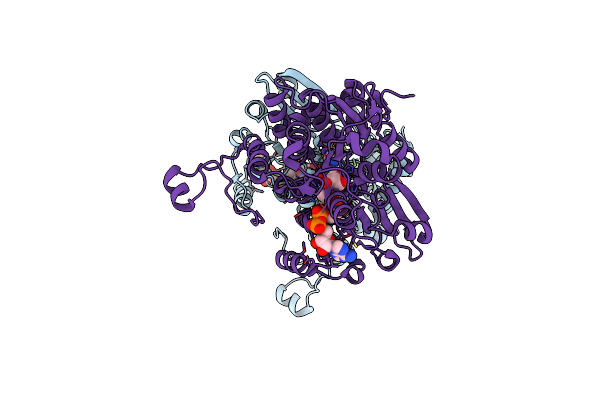 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Mus Musculus In Complex With Inosine
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: NAD, NOS, NA |
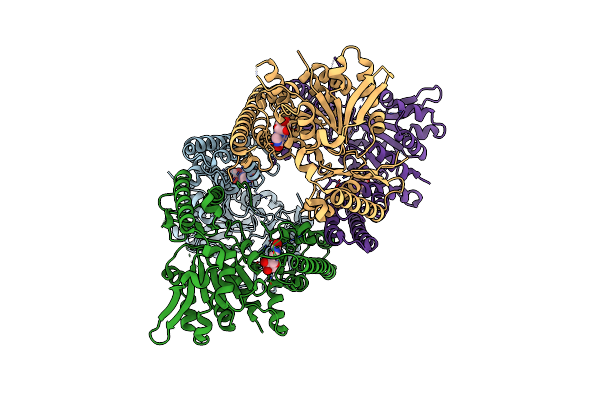 |
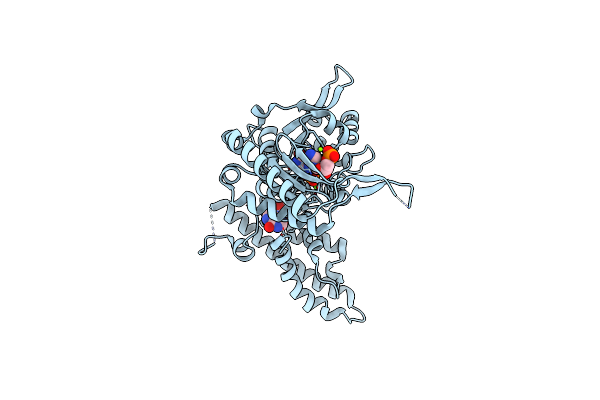
Structure And Allosteric Regulation Of The Inosine 5'-Monophosphate-Specific Phosphatase Isn1 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: NOS |
 |
Structure And Allosteric Regulation Of The Inosine 5'-Monophosphate-Specific Phosphatase Isn1 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-03-06 Classification: HYDROLASE Ligands: NOS, PO4, IMP, MG |
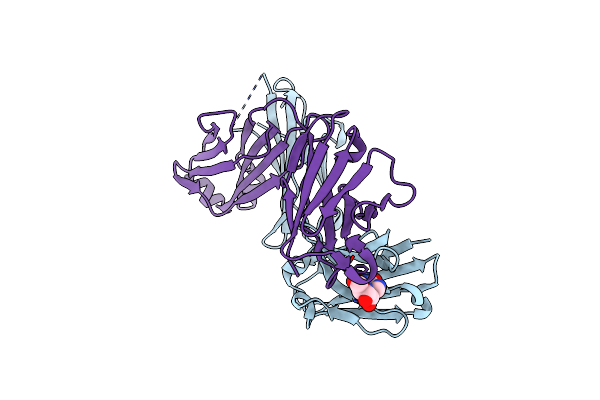 |
Structure Of A Mouse Igg Antibody Fragment That Binds Inosine, An Rna Modification
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-02-07 Classification: IMMUNE SYSTEM Ligands: NOS |
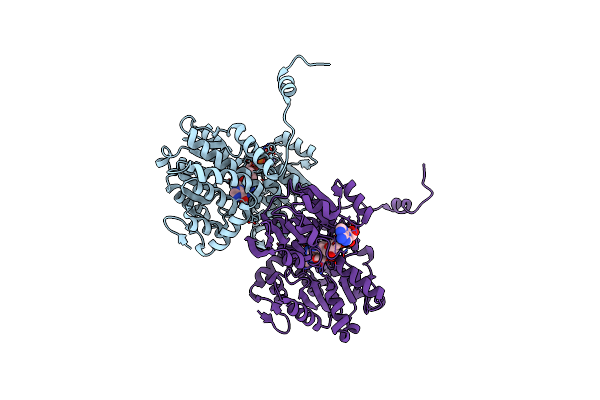 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Pyrococcus Furiosus In Complex With Inosine
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: NAD, NOS |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Methanococcus Maripaludis In Complex With Inosine
Organism: Methanococcus maripaludis
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: NOS, NAD, PG4 |
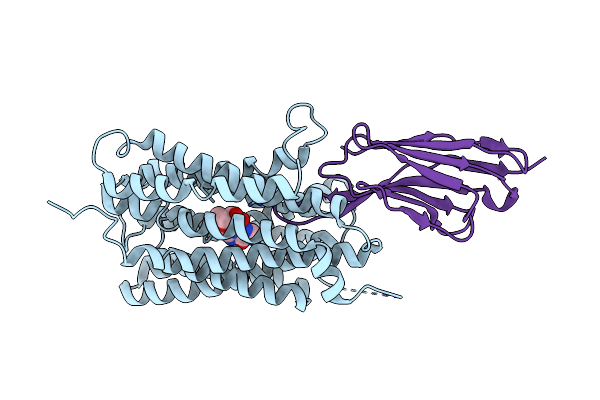 |
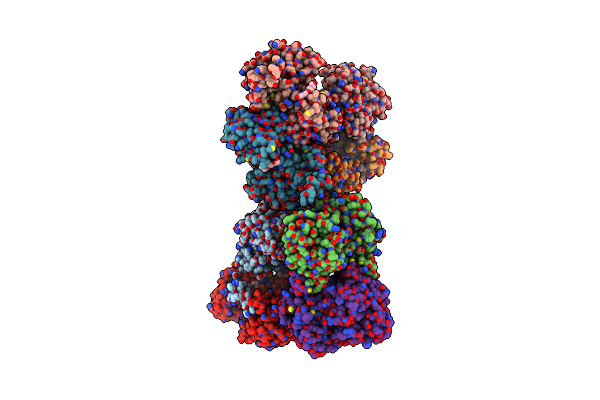
Organism: Plasmodium falciparum, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: TRANSPORT PROTEIN Ligands: NOS |
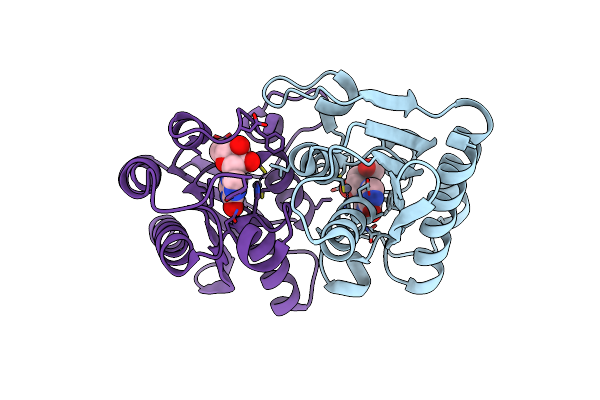 |
The Structure Of The Arabidopsis Thaliana Guanosine Deaminase Complexed With Inosine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-10-27 Classification: HYDROLASE Ligands: ZN, NOS |
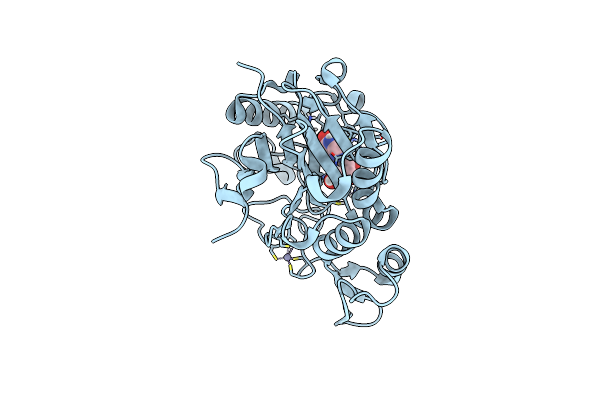 |
Crystal Structure Of Ylmd From Geobacillus Stearothermophilus In Complex With Inosine
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2020-01-29 Classification: HYDROLASE Ligands: NOS, ZN |
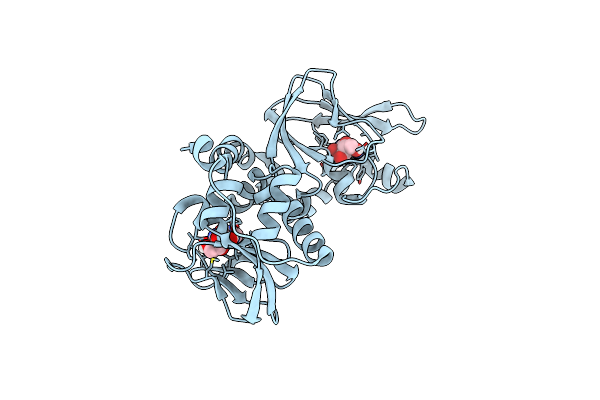 |
Regulatory Subunit Of A Camp-Independent Protein Kinase A From Trypanosoma Cruzi At 1.4 A Resolution In Complex With Inosine
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-11-13 Classification: SIGNALING PROTEIN Ligands: NOS |
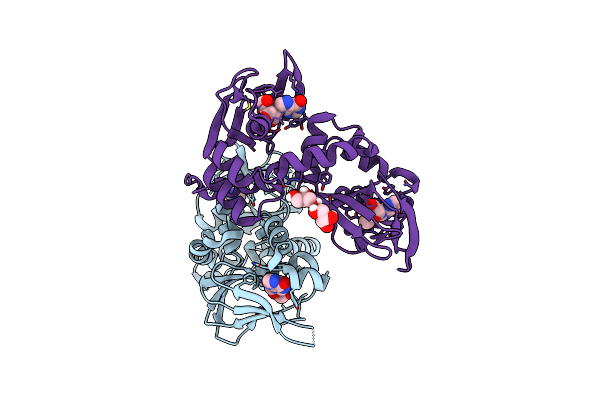 |
Regulatory Subunit Of A Camp-Independent Protein Kinase A From Trypanosoma Brucei At 2.1 Angstrom Resolution
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2019-08-14 Classification: SIGNALING PROTEIN Ligands: NOS, GOL |
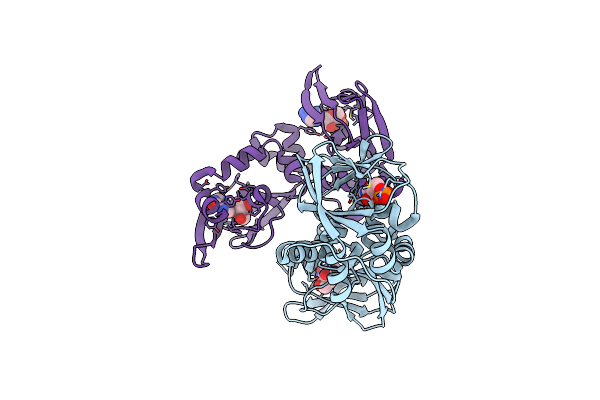 |
Regulatory Subunit Of A Camp-Independent Protein Kinase A From Trypanosoma Brucei: E311A, T318R, V319A Mutant Bound To Camp In The A Site
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2019-08-14 Classification: SIGNALING PROTEIN Ligands: CMP, NOS |
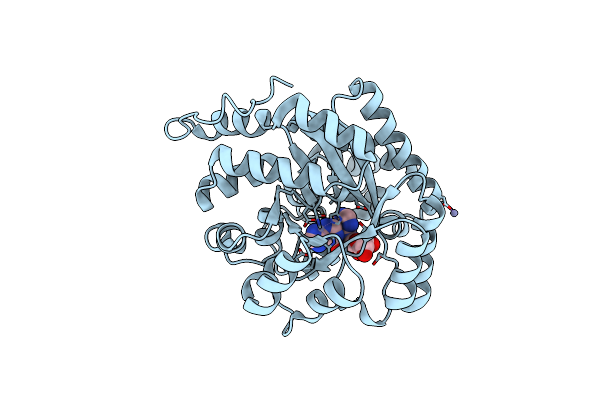 |
Crystal Structure Of Plasmodium Falciparum Adenosine Deaminase C27Q+L227I Mutant Co-Complexed With Zn Ion, Hypoxanthine And Inosine
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2019-05-01 Classification: HYDROLASE Ligands: NOS, HPA, ZN |
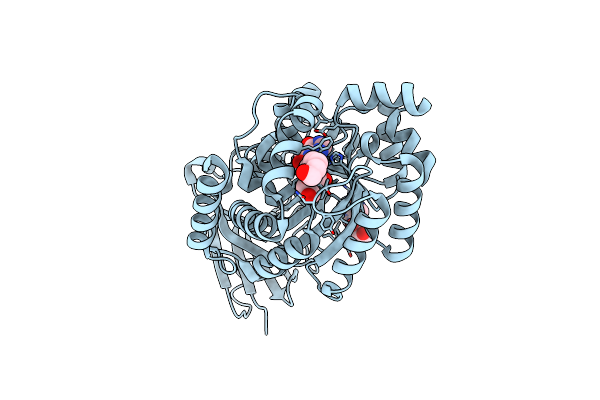 |
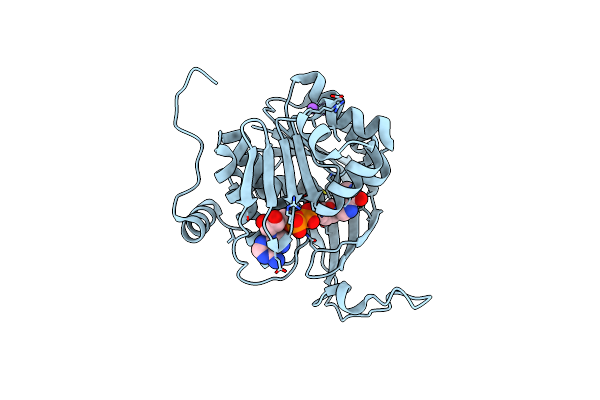
Crystal Structure Of An Adenosine Deaminase Homolog From Chromobacterium Violaceum (Target Nysgrc-019589) With Bound Inosine.
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-06-06 Classification: HYDROLASE Ligands: NOS, CL, PG4 |
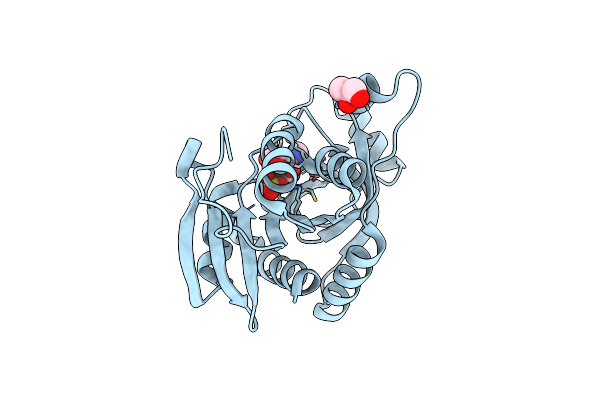 |
Crystal Structure Of Adenosine Phosphorylase From Bacillus Cereus Complexed With Inosine
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2012-02-29 Classification: TRANSFERASE Ligands: NOS, SO4, GOL |
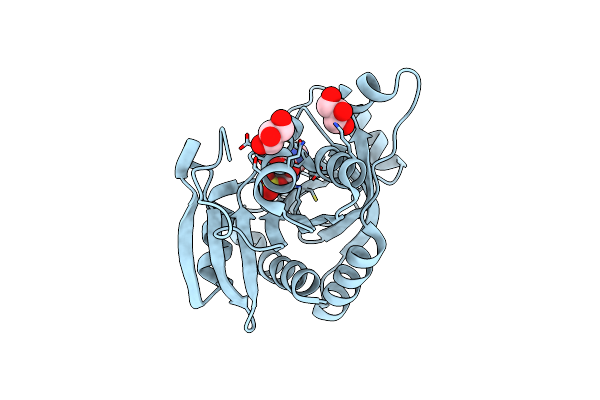 |
Crystal Structure Of Bacillus Cereus Adenosine Phosphorylase D204N Mutant Complexed With Inosine
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-02-29 Classification: TRANSFERASE Ligands: NOS, SO4, GOL |
 |
Structure Of Burkholderia Thailandensis Nucleoside Kinase (Bthnk) In Complex With Adp-Inosine
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-11-02 Classification: TRANSFERASE Ligands: NOS, ADP, NA |
 |
Structure Of Burkholderia Thailandensis Nucleoside Kinase (Bthnk) In Complex With Inosine
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-11-02 Classification: TRANSFERASE Ligands: NOS, ACT, P33, NA |

