Search Count: 953
 |
Organism: Parengyodontium album
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: EU, NO3 |
 |
Crystal Structure Of Pleurocybella Porrigens Lectin (Ppl) In Complex With Galnac
Organism: Pleurocybella porrigens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: SUGAR BINDING PROTEIN Ligands: A2G, NO3 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: CYTOKINE Ligands: NO3 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: METAL BINDING PROTEIN Ligands: NI, NO3 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: ZN, NO3 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: FLUORESCENT PROTEIN Ligands: NO3, SO4 |
 |
Crystal Structure Of Molybdenum Bispyranopterin Guanine Dinucleotide Formate Dehydrogenases Force1 From Bacillus Subtilis
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: MYR, GOL, LHG, FES, SF4, MGD, 4MO, MQ7, H2S, MLI, NO3, NA |
 |
Organism: Homo sapiens, Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TRANSPORT PROTEIN Ligands: GTP, MG, GOL, NA, NO3 |
 |
X-Ray Structure Of Human Acetylcholinesterase (Hache) In Complex With Bis-Oxime Reactivator Lg-1922
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: A1CD5, NO3, GOL |
 |
X-Ray Structure Of Paraoxon (Pox)-Inhibited Human Acetylcholinesterase (Hache) In Complex With Bis-Oxime Reactivator Lg-1922
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: DEP, GOL, NO3, A1CDA, DMS |
 |
Crystal Structure Of Isoform Chitin Binding Protein From Iberis Umbellata L.
Organism: Iberis umbellata
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: PLANT PROTEIN Ligands: SO4, NA, NO3, CL, ACT |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Organism: Parengyodontium album
Method: ELECTRON CRYSTALLOGRAPHY Resolution:1.30 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: CA, NO3 |
 |
Organism: Parengyodontium album
Method: ELECTRON CRYSTALLOGRAPHY Resolution:1.30 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: CA, NO3 |
 |
Crystal Structure Of Lmrr Variant V15Ay-Rnyw With Val15 Replaced By 3-Aminotyrosine And Evolved As Friedel-Crafts Alkylase
Organism: Lactococcus cremoris subsp. cremoris mg1363
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: DNA BINDING PROTEIN Ligands: NO3 |
 |
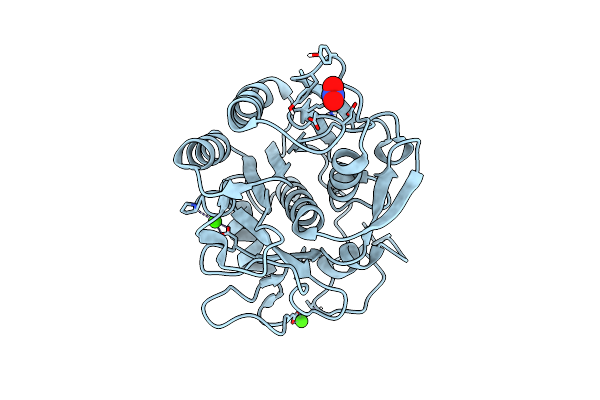
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: HYDROLASE Ligands: NO3, EDO |
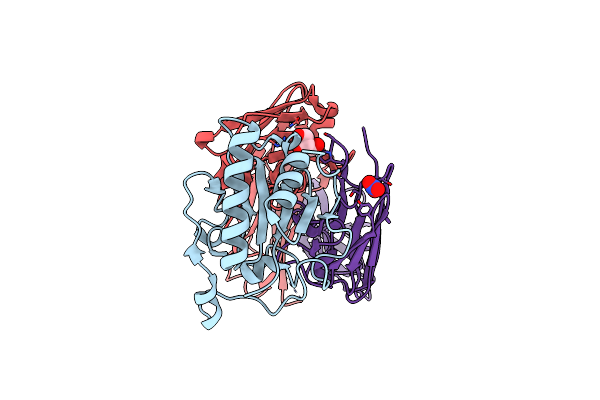 |
Crystal Structure Of Sonic Hedgehog In Complex With Antibody 5E1 Without Metals
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-04-16 Classification: IMMUNE SYSTEM Ligands: GOL, NO3 |
 |
Crystal Structure Of The Light-Driven Proton Pump Heimdallarchaeial Rhodopsin Heimdallr1
Organism: Candidatus heimdallarchaeota archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: PROTON TRANSPORT Ligands: RET, OLC, NO3 |
 |
Organism: Parengyodontium album
Method: ELECTRON CRYSTALLOGRAPHY Release Date: 2025-03-19 Classification: HYDROLASE Ligands: CA, NO3 |

