Search Count: 53
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: TRANSFERASE Ligands: NMN, GOL, PEG, EDO |
 |
Structural Insights Into The Nmn Complex Of Nicotinate Nucleotide Adenylyltransferase From Enterococcus Faecium Via Co-Crystallization Studies
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2024-07-17 Classification: TRANSFERASE Ligands: NMN, PGE, PEG, PO4, MG |
 |
Organism: Enterococcus faecalis (strain atcc 700802 / v583)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-07-10 Classification: LIGASE Ligands: NMN |
 |
Structure Of The Non-Canonical Ctlh E3 Substrate Receptor Wdr26 Bound To Nmnat1 Substrate
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: LIGASE Ligands: NMN, ZN |
 |
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-02-14 Classification: TRANSFERASE Ligands: NMN, PO4 |
 |
Complex Structure Of A Glucose 6-Phosphate Dehydrogenase From Zymomonas Mobilis
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2023-04-12 Classification: OXIDOREDUCTASE Ligands: NMN |
 |
Organism: Streptococcus parasanguinis
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2023-03-22 Classification: RNA Ligands: MG, NMN |
 |
Organism: Streptococcus parasanguinis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-03-22 Classification: RNA Ligands: NMN, BA |
 |
Organism: Streptococcus parasanguinis
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2023-03-22 Classification: RNA Ligands: NAD, NMN |
 |
Organism: Homo sapiens, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: HYDROLASE Ligands: NMN |
 |
Organism: Homo sapiens, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: HYDROLASE Ligands: NMN |
 |
Organism: Streptococcus sp., Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-01-18 Classification: PROTEIN/RNA Ligands: NMN, MG |
 |
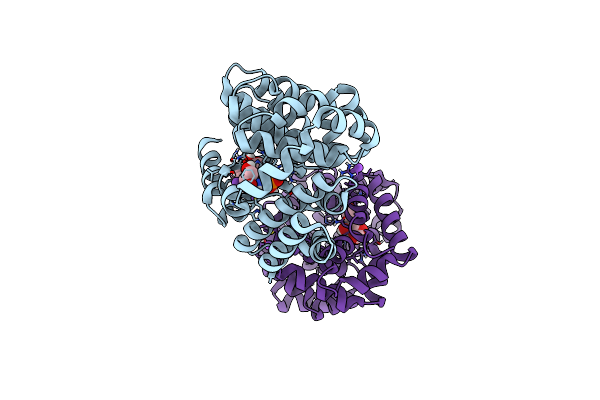
Cryo-Em Structure Of Activated Human Sarm1 In Complex With Nmn And 1Ad (Arm And Sam Domains)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-23 Classification: HYDROLASE Ligands: NMN |
 |
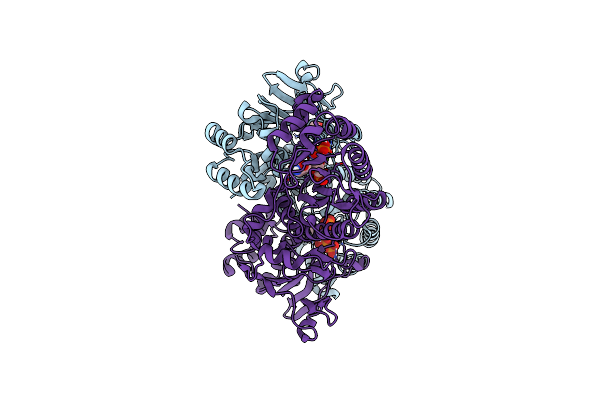
Crystal Structure Of The Arm Domain From Drosophila Sarm1 In Complex With Nmn
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-03-10 Classification: HYDROLASE Ligands: NMN, EDO, NA |
 |
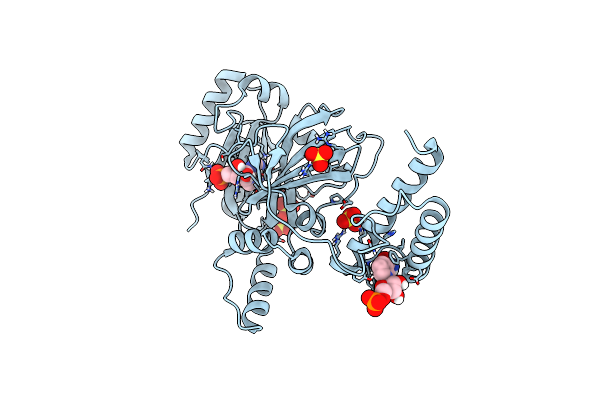
Human Nampt Deletion Mutant In Complex With Nicotinamide Mononucleotide, Pyrophosphate, And Mg2+
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: GOL, MG, NMN, PPV |
 |
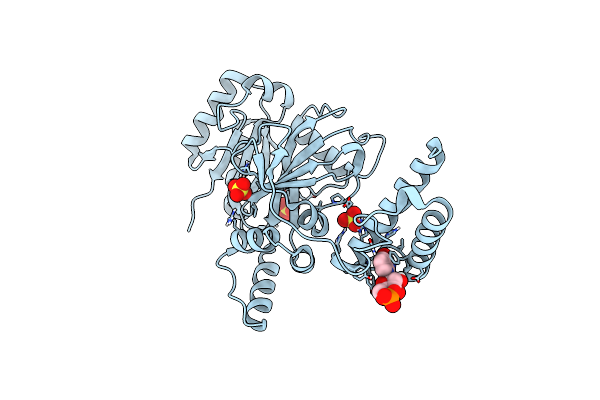
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-08-26 Classification: LIGASE Ligands: AMP, NMN, SO4 |
 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-08-26 Classification: LIGASE Ligands: NMN, SO4 |
 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-22 Classification: LIGASE Ligands: NMN, AMP, SO4 |
 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-01 Classification: DNA BINDING PROTEIN Ligands: AMP, NMN, SO4 |
 |
Organism: Lactococcus lactis subsp. cremoris
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-07-10 Classification: TRANSPORT PROTEIN Ligands: NMN, SO4, MG |

